6 Traits, 5 tissues, eQTL + sQTL + stQTL – compute ukbb LD, eqtl, sqtl from predictdb; stQTL from Munro et al; post-processing
XSun
2024-12-03
Last updated: 2024-12-06
Checks: 6 1
Knit directory: multigroup_ctwas_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of
the R Markdown file created these results, you’ll want to first commit
it to the Git repo. If you’re still working on the analysis, you can
ignore this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20231112) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 984b87f. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Unstaged changes:
Modified: analysis/multi_group_6traits_15weights_ess_postprocessing.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/multi_group_6traits_15weights_ess_postprocessing.Rmd)
and HTML
(docs/multi_group_6traits_15weights_ess_postprocessing.html)
files. If you’ve configured a remote Git repository (see
?wflow_git_remote), click on the hyperlinks in the table
below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 984b87f | XSun | 2024-12-06 | update |
| html | 984b87f | XSun | 2024-12-06 | update |
| Rmd | 4885dec | XSun | 2024-12-05 | update |
| html | 4885dec | XSun | 2024-12-05 | update |
| Rmd | 32ca054 | XSun | 2024-12-03 | update |
Introduction
We compare the post-processed results with the original results here: https://sq-96.github.io/multigroup_ctwas_analysis/multi_group_6traits_15weights_ess.html
library(ctwas)
library(EnsDb.Hsapiens.v86)
library(ggplot2)
library(gridExtra)
library(dplyr)
ens_db <- EnsDb.Hsapiens.v86
mapping_predictdb <- readRDS("/project2/xinhe/shared_data/multigroup_ctwas/weights/mapping_files/PredictDB_mapping.RDS")
mapping_munro <- readRDS("/project2/xinhe/shared_data/multigroup_ctwas/weights/mapping_files/Munro_mapping.RDS")
mapping_two <- rbind(mapping_predictdb,mapping_munro)
compute_pip_per_cs <- function(combined_data, susie_data) {
# Initialize an empty list to store results
details <- list()
# Iterate over each unique gene name in the combined data
unique_genes <- unique(combined_data$gene_name)
for (genename in unique_genes) {
# dplyr::filter susie data for the current gene
susie_alpha_res_multi_per_gene <- susie_data %>%
dplyr::filter(gene_name == genename)
# Get all unique credible sets for the current gene
cs_all <- unique(susie_alpha_res_multi_per_gene$susie_set[susie_alpha_res_multi_per_gene$in_cs])
if (length(cs_all) > 1) {
# dplyr::filter complete cases and those in credible sets
susie_alpha_res_multi_per_gene <- susie_alpha_res_multi_per_gene %>%
dplyr::filter(complete.cases(cs), in_cs)
# Summarize the data
summed_alpha_with_details <- susie_alpha_res_multi_per_gene %>%
group_by(susie_set) %>%
summarise(
total_susie_alpha = round(sum(susie_alpha, na.rm = TRUE), digits = 3),
num_molecular_traits = n(),
ids_pip = paste0(id, "(", round(susie_alpha, digits = 3), ")", collapse = ", ")
)
# Add gene name to the summarized data
summed_alpha_with_details$gene_name <- genename
# Append the result to the details list
details[[length(details) + 1]] <- summed_alpha_with_details
}
}
# Combine all results into a single data frame
final_details <- bind_rows(details)
if(nrow(final_details) > 0){
final_details <- final_details[,c("gene_name","susie_set","total_susie_alpha","num_molecular_traits","ids_pip")]
colnames(final_details) <- c("gene_name","CS","total_PIP_CS","num_molecular_traits_CS","ids_pip_CS")
}
return(final_details)
}aFib-ebi-a-GCST006414
trait <- "aFib-ebi-a-GCST006414"
results_dir_origin <- paste0("/project/xinhe/xsun/multi_group_ctwas/11.multi_group_1008/results/",trait,"/")
ctwas_res_origin <- readRDS(paste0(results_dir_origin,trait,".ctwas.res.RDS"))
finemap_res_origin <- ctwas_res_origin$finemap_resRegion merge
load(paste0("/project/xinhe/xsun/multi_group_ctwas/11.multi_group_1008/post_process_rm_ld/rm_",trait,".rdata"))
finemap_res_rm <- res_regionmerge$finemap_res
finemap_res_rm_boundary_genes <- finemap_res_rm[finemap_res_rm$id %in%selected_boundary_genes$id,]
finemap_res_rm_boundary_genes_pip <- finemap_res_rm_boundary_genes[,c("id","susie_pip","cs")]
finemap_res_origin_boundary_genes <- finemap_res_origin[finemap_res_origin$id %in%selected_boundary_genes$id,]
finemap_res_origin_boundary_genes_pip <- finemap_res_origin_boundary_genes[,c("id","susie_pip","cs")]
finemap_res_compare_regionmerge <- merge(finemap_res_origin_boundary_genes_pip,finemap_res_rm_boundary_genes_pip, by = "id")
colnames(finemap_res_compare_regionmerge) <- c("id","susie_pip_origin","cs_origin","susie_pip_reginmerge","cs_reginmerge")
DT::datatable(finemap_res_compare_regionmerge,caption = htmltools::tags$caption( style = 'caption-side: left; text-align: left; color:black; font-size:150% ;','Selected boundary genes (susie_pip > 0.5)'),options = list(pageLength = 10) )LD-mismatch
load(paste0("/project/xinhe/xsun/multi_group_ctwas/11.multi_group_1008/post_process_rm_ld/rmld_",trait,".rdata"))
sprintf("The number of problematic regions = %s", length(problematic_region_ids))[1] "The number of problematic regions = 4"sprintf("The number of problematic genes = %s", length(problematic_genes))[1] "The number of problematic genes = 4"sprintf("The number of problematic snps = %s", length(res_ldmismatch$problematic_snps))[1] "The number of problematic snps = 1712"sprintf("The number of flipped snps = %s", length(res_ldmismatch$flipped_snps))[1] "The number of flipped snps = 22"finemap_rm_res_problematic_region <- finemap_res_rm[finemap_res_rm$id %in% problematic_genes,]
finemap_noLD_res_problematic_region <- res_rmld$finemap_res
merge_regionmerge_nold <- merge(finemap_rm_res_problematic_region,finemap_noLD_res_problematic_region, by = "id")
merge_regionmerge_nold <- merge_regionmerge_nold[,c("id","susie_pip.x","susie_pip.y")]
colnames(merge_regionmerge_nold) <- c("id","susie_pip_after_regionmerge","susie_pip_ld-mismatch-fixed")
DT::datatable(merge_regionmerge_nold,caption = htmltools::tags$caption(style = 'caption-side: topleft; text-align = left; color:black;','PIP after region merge and fixed PIP for problematic genes'),options = list(pageLength = 5) )finemap_res_origin <- ctwas_res_origin$finemap_res
finemap_res_origin_gene <- finemap_res_origin[finemap_res_origin$type != "SNP",]
p1 <- ggplot(data = finemap_res_origin_gene, aes(x= abs(z), y= susie_pip)) +
geom_point() +
ggtitle("Original ctwas results") +
theme_minimal()
finemap_res_rm_gene <- finemap_res_rm[finemap_res_rm$type != "SNP",]
p2 <- ggplot(data = finemap_res_rm_gene, aes(x= abs(z), y= susie_pip)) +
geom_point() +
ggtitle("After region merge") +
theme_minimal()
finemap_noLD_res_problematic_region_gene <- finemap_noLD_res_problematic_region[finemap_noLD_res_problematic_region$type !="SNP",]
p3 <- ggplot(data = finemap_noLD_res_problematic_region_gene, aes(x= abs(z), y= susie_pip)) +
geom_point() +
ggtitle("After LD mismatch fixed") +
theme_minimal()
grid.arrange(p1,p2,p3, ncol = 3)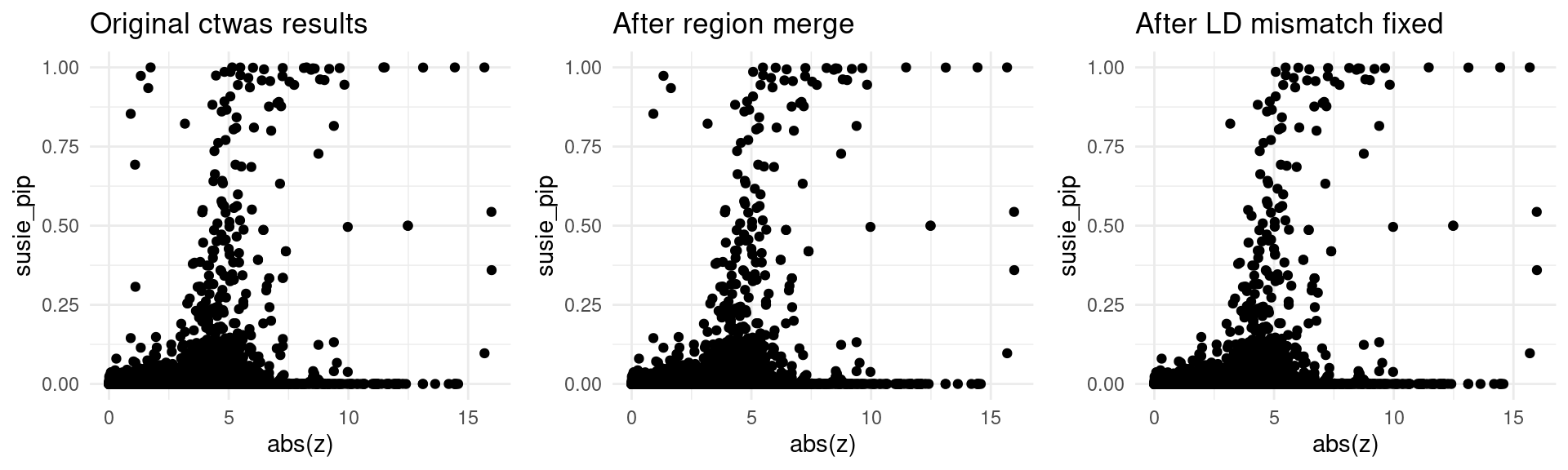
| Version | Author | Date |
|---|---|---|
| 4885dec | XSun | 2024-12-05 |
Comparing results without post processing
susie_alpha_res_origin <- ctwas_res_origin$susie_alpha_res
susie_alpha_res_origin <- anno_susie_alpha_res(susie_alpha_res_origin,
mapping_table = mapping_two,
map_by = "molecular_id",
drop_unmapped = TRUE)2024-12-06 13:21:56 INFO::Annotating susie alpha result ...
2024-12-06 13:21:56 INFO::Map molecular traits to genes
2024-12-06 13:21:57 INFO::Split PIPs for molecular traits mapped to multiple genescombined_pip_by_type_cs_origin <- combine_gene_pips(susie_alpha_res_origin,
group_by = "gene_name",
by = "type",
method = "combine_cs",
filter_cs = TRUE,
include_cs_id = T)
combined_pip_by_type_sig_origin <- combined_pip_by_type_cs_origin[combined_pip_by_type_cs_origin$combined_pip > 0.8,]
susie_alpha_res_rm <- res_regionmerge$susie_alpha_res
susie_alpha_res_rm <- anno_susie_alpha_res(susie_alpha_res_rm,
mapping_table = mapping_two,
map_by = "molecular_id",
drop_unmapped = TRUE)2024-12-06 13:22:03 INFO::Annotating susie alpha result ...
2024-12-06 13:22:03 INFO::Map molecular traits to genes
2024-12-06 13:22:03 INFO::Split PIPs for molecular traits mapped to multiple genescombined_pip_by_type_cs_rm <- combine_gene_pips(susie_alpha_res_rm,
group_by = "gene_name",
by = "type",
method = "combine_cs",
filter_cs = TRUE,
include_cs_id = T)
combined_pip_by_type_sig_rm <- combined_pip_by_type_cs_rm[combined_pip_by_type_cs_rm$combined_pip > 0.8,]
susie_alpha_res_rmld <- res_rmld$susie_alpha_res
susie_alpha_res_rmld <- anno_susie_alpha_res(susie_alpha_res_rmld,
mapping_table = mapping_two,
map_by = "molecular_id",
drop_unmapped = TRUE)2024-12-06 13:22:07 INFO::Annotating susie alpha result ...
2024-12-06 13:22:07 INFO::Map molecular traits to genes
2024-12-06 13:22:08 INFO::Split PIPs for molecular traits mapped to multiple genescombined_pip_by_type_cs_rmld <- combine_gene_pips(susie_alpha_res_rmld,
group_by = "gene_name",
by = "type",
method = "combine_cs",
filter_cs = TRUE,
include_cs_id = T)
combined_pip_by_type_sig_rmld <- combined_pip_by_type_cs_rmld[combined_pip_by_type_cs_rmld$combined_pip > 0.8,]
origin_unique <- combined_pip_by_type_sig_origin[!combined_pip_by_type_sig_origin$gene_name %in% combined_pip_by_type_sig_rmld$gene_name,]
origin_unique <- merge(origin_unique, combined_pip_by_type_cs_rm, by = "gene_name", all.x = TRUE)
# Then merge the result with combined_pip_by_type_cs_rmld
origin_unique <- merge(origin_unique, combined_pip_by_type_cs_rmld, by = "gene_name", all.x = TRUE)
origin_unique_show <- origin_unique[,c("gene_name","combined_pip.x","combined_pip.y","combined_pip","eQTL_pip.x","eQTL_pip.y","eQTL_pip","sQTL_pip.x","sQTL_pip.y","sQTL_pip","stQTL_pip.x","stQTL_pip.x","stQTL_pip","combined_cs_id.x","combined_cs_id.y","combined_cs_id")]
colnames(origin_unique_show) <- c("gene_name","combined_pip_origin","combined_pip_rm","combined_pip_rmld","eQTL_pip_origin","eQTL_pip_rm","eQTL_pip_rmld","sQTL_pip_origin","sQTL_pip_rm","sQTL_pip_rmld","stQTL_pip_origin","stQTL_pip_origin","stQTL_pip_rmld","combined_cs_id_origin","combined_cs_id_rm","combined_cs_id_rmld")
DT::datatable(origin_unique_show,caption = htmltools::tags$caption(style = 'caption-side: top; text-align: left; color: black;','Genes no longer significant after post processing'),options = list(pageLength = 5))Exploring allelic heterogeneity
pip_per_cs <- compute_pip_per_cs(combined_pip_by_type_sig_rmld, susie_alpha_res_rmld)
sprintf("Number of genes having allelic heterogeneity = %s",length(unique(pip_per_cs$gene_name)))[1] "Number of genes having allelic heterogeneity = 4"if(nrow(pip_per_cs) > 0) {
DT::datatable(pip_per_cs,caption = htmltools::tags$caption( style = 'caption-side: topleft; text-align = left; color:black;','PIP per CS'),options = list(pageLength = 5) )
}LDL-ukb-d-30780_irnt
trait <- "LDL-ukb-d-30780_irnt"
results_dir_origin <- paste0("/project/xinhe/xsun/multi_group_ctwas/11.multi_group_1008/results/",trait,"/")
ctwas_res_origin <- readRDS(paste0(results_dir_origin,trait,".ctwas.res.RDS"))
finemap_res_origin <- ctwas_res_origin$finemap_resRegion merge
load(paste0("/project/xinhe/xsun/multi_group_ctwas/11.multi_group_1008/post_process_rm_ld/rm_",trait,".rdata"))
finemap_res_rm <- res_regionmerge$finemap_res
finemap_res_rm_boundary_genes <- finemap_res_rm[finemap_res_rm$id %in%selected_boundary_genes$id,]
finemap_res_rm_boundary_genes_pip <- finemap_res_rm_boundary_genes[,c("id","susie_pip","cs")]
finemap_res_origin_boundary_genes <- finemap_res_origin[finemap_res_origin$id %in%selected_boundary_genes$id,]
finemap_res_origin_boundary_genes_pip <- finemap_res_origin_boundary_genes[,c("id","susie_pip","cs")]
finemap_res_compare_regionmerge <- merge(finemap_res_origin_boundary_genes_pip,finemap_res_rm_boundary_genes_pip, by = "id")
colnames(finemap_res_compare_regionmerge) <- c("id","susie_pip_origin","cs_origin","susie_pip_reginmerge","cs_reginmerge")
DT::datatable(finemap_res_compare_regionmerge,caption = htmltools::tags$caption( style = 'caption-side: left; text-align: left; color:black; font-size:150% ;','Selected boundary genes (susie_pip > 0.5)'),options = list(pageLength = 10) )LD-mismatch
load(paste0("/project/xinhe/xsun/multi_group_ctwas/11.multi_group_1008/post_process_rm_ld/rmld_",trait,".rdata"))
sprintf("The number of problematic regions = %s", length(problematic_region_ids))[1] "The number of problematic regions = 2"sprintf("The number of problematic genes = %s", length(problematic_genes))[1] "The number of problematic genes = 5"sprintf("The number of problematic snps = %s", length(res_ldmismatch$problematic_snps))[1] "The number of problematic snps = 325"sprintf("The number of flipped snps = %s", length(res_ldmismatch$flipped_snps))[1] "The number of flipped snps = 8"finemap_rm_res_problematic_region <- finemap_res_rm[finemap_res_rm$id %in% problematic_genes,]
finemap_noLD_res_problematic_region <- res_rmld$finemap_res
merge_regionmerge_nold <- merge(finemap_rm_res_problematic_region,finemap_noLD_res_problematic_region, by = "id")
merge_regionmerge_nold <- merge_regionmerge_nold[,c("id","susie_pip.x","susie_pip.y")]
colnames(merge_regionmerge_nold) <- c("id","susie_pip_after_regionmerge","susie_pip_ld-mismatch-fixed")
DT::datatable(merge_regionmerge_nold,caption = htmltools::tags$caption(style = 'caption-side: topleft; text-align = left; color:black;','PIP after region merge and fixed PIP for problematic genes'),options = list(pageLength = 5) )finemap_res_origin <- ctwas_res_origin$finemap_res
finemap_res_origin_gene <- finemap_res_origin[finemap_res_origin$type != "SNP",]
p1 <- ggplot(data = finemap_res_origin_gene, aes(x= abs(z), y= susie_pip)) +
geom_point() +
ggtitle("Original ctwas results") +
theme_minimal()
finemap_res_rm_gene <- finemap_res_rm[finemap_res_rm$type != "SNP",]
p2 <- ggplot(data = finemap_res_rm_gene, aes(x= abs(z), y= susie_pip)) +
geom_point() +
ggtitle("After region merge") +
theme_minimal()
finemap_noLD_res_problematic_region_gene <- finemap_noLD_res_problematic_region[finemap_noLD_res_problematic_region$type !="SNP",]
p3 <- ggplot(data = finemap_noLD_res_problematic_region_gene, aes(x= abs(z), y= susie_pip)) +
geom_point() +
ggtitle("After LD mismatch fixed") +
theme_minimal()
grid.arrange(p1,p2,p3, ncol = 3)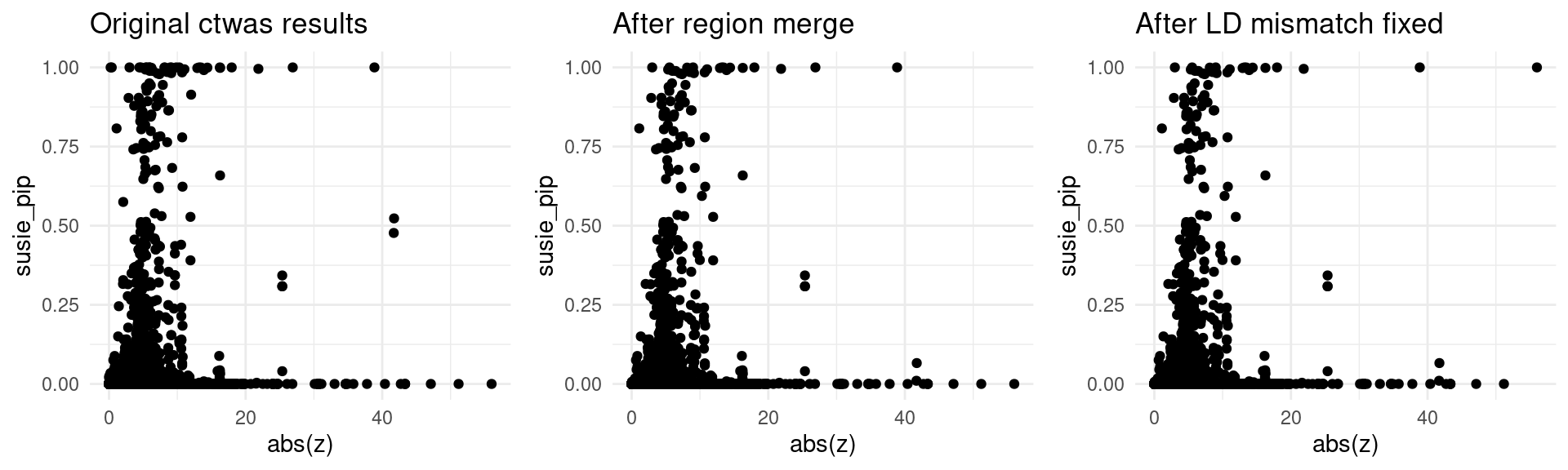
| Version | Author | Date |
|---|---|---|
| 4885dec | XSun | 2024-12-05 |
Comparing results without post processing
susie_alpha_res_origin <- ctwas_res_origin$susie_alpha_res
susie_alpha_res_origin <- anno_susie_alpha_res(susie_alpha_res_origin,
mapping_table = mapping_two,
map_by = "molecular_id",
drop_unmapped = TRUE)2024-12-06 13:22:55 INFO::Annotating susie alpha result ...
2024-12-06 13:22:55 INFO::Map molecular traits to genes
2024-12-06 13:22:56 INFO::Split PIPs for molecular traits mapped to multiple genescombined_pip_by_type_cs_origin <- combine_gene_pips(susie_alpha_res_origin,
group_by = "gene_name",
by = "type",
method = "combine_cs",
filter_cs = TRUE,
include_cs_id = T)
combined_pip_by_type_sig_origin <- combined_pip_by_type_cs_origin[combined_pip_by_type_cs_origin$combined_pip > 0.8,]
susie_alpha_res_rm <- res_regionmerge$susie_alpha_res
susie_alpha_res_rm <- anno_susie_alpha_res(susie_alpha_res_rm,
mapping_table = mapping_two,
map_by = "molecular_id",
drop_unmapped = TRUE)2024-12-06 13:23:00 INFO::Annotating susie alpha result ...
2024-12-06 13:23:00 INFO::Map molecular traits to genes
2024-12-06 13:23:01 INFO::Split PIPs for molecular traits mapped to multiple genescombined_pip_by_type_cs_rm <- combine_gene_pips(susie_alpha_res_rm,
group_by = "gene_name",
by = "type",
method = "combine_cs",
filter_cs = TRUE,
include_cs_id = T)
combined_pip_by_type_sig_rm <- combined_pip_by_type_cs_rm[combined_pip_by_type_cs_rm$combined_pip > 0.8,]
susie_alpha_res_rmld <- res_rmld$susie_alpha_res
susie_alpha_res_rmld <- anno_susie_alpha_res(susie_alpha_res_rmld,
mapping_table = mapping_two,
map_by = "molecular_id",
drop_unmapped = TRUE)2024-12-06 13:23:07 INFO::Annotating susie alpha result ...
2024-12-06 13:23:07 INFO::Map molecular traits to genes
2024-12-06 13:23:07 INFO::Split PIPs for molecular traits mapped to multiple genescombined_pip_by_type_cs_rmld <- combine_gene_pips(susie_alpha_res_rmld,
group_by = "gene_name",
by = "type",
method = "combine_cs",
filter_cs = TRUE,
include_cs_id = T)
combined_pip_by_type_sig_rmld <- combined_pip_by_type_cs_rmld[combined_pip_by_type_cs_rmld$combined_pip > 0.8,]
origin_unique <- combined_pip_by_type_sig_origin[!combined_pip_by_type_sig_origin$gene_name %in% combined_pip_by_type_sig_rmld$gene_name,]
origin_unique <- merge(origin_unique, combined_pip_by_type_cs_rm, by = "gene_name", all.x = TRUE)
# Then merge the result with combined_pip_by_type_cs_rmld
origin_unique <- merge(origin_unique, combined_pip_by_type_cs_rmld, by = "gene_name", all.x = TRUE)
origin_unique_show <- origin_unique[,c("gene_name","combined_pip.x","combined_pip.y","combined_pip","eQTL_pip.x","eQTL_pip.y","eQTL_pip","sQTL_pip.x","sQTL_pip.y","sQTL_pip","stQTL_pip.x","stQTL_pip.x","stQTL_pip","combined_cs_id.x","combined_cs_id.y","combined_cs_id")]
colnames(origin_unique_show) <- c("gene_name","combined_pip_origin","combined_pip_rm","combined_pip_rmld","eQTL_pip_origin","eQTL_pip_rm","eQTL_pip_rmld","sQTL_pip_origin","sQTL_pip_rm","sQTL_pip_rmld","stQTL_pip_origin","stQTL_pip_origin","stQTL_pip_rmld","combined_cs_id_origin","combined_cs_id_rm","combined_cs_id_rmld")
DT::datatable(origin_unique_show,caption = htmltools::tags$caption(style = 'caption-side: top; text-align: left; color: black;','Genes no longer significant after post processing'),options = list(pageLength = 5))Exploring allelic heterogeneity
pip_per_cs <- compute_pip_per_cs(combined_pip_by_type_sig_rmld, susie_alpha_res_rmld)
sprintf("Number of genes having allelic heterogeneity = %s",length(unique(pip_per_cs$gene_name)))[1] "Number of genes having allelic heterogeneity = 5"if(nrow(pip_per_cs) > 0) {
DT::datatable(pip_per_cs,caption = htmltools::tags$caption( style = 'caption-side: topleft; text-align = left; color:black;','PIP per CS'),options = list(pageLength = 5) )
}IBD-ebi-a-GCST004131
trait <- "IBD-ebi-a-GCST004131"
results_dir_origin <- paste0("/project/xinhe/xsun/multi_group_ctwas/11.multi_group_1008/results/",trait,"/")
ctwas_res_origin <- readRDS(paste0(results_dir_origin,trait,".ctwas.res.RDS"))
finemap_res_origin <- ctwas_res_origin$finemap_resRegion merge
load(paste0("/project/xinhe/xsun/multi_group_ctwas/11.multi_group_1008/post_process_rm_ld/rm_",trait,".rdata"))
finemap_res_rm <- res_regionmerge$finemap_res
finemap_res_rm_boundary_genes <- finemap_res_rm[finemap_res_rm$id %in%selected_boundary_genes$id,]
finemap_res_rm_boundary_genes_pip <- finemap_res_rm_boundary_genes[,c("id","susie_pip","cs")]
finemap_res_origin_boundary_genes <- finemap_res_origin[finemap_res_origin$id %in%selected_boundary_genes$id,]
finemap_res_origin_boundary_genes_pip <- finemap_res_origin_boundary_genes[,c("id","susie_pip","cs")]
finemap_res_compare_regionmerge <- merge(finemap_res_origin_boundary_genes_pip,finemap_res_rm_boundary_genes_pip, by = "id")
colnames(finemap_res_compare_regionmerge) <- c("id","susie_pip_origin","cs_origin","susie_pip_reginmerge","cs_reginmerge")
DT::datatable(finemap_res_compare_regionmerge,caption = htmltools::tags$caption( style = 'caption-side: left; text-align: left; color:black; font-size:150% ;','Selected boundary genes (susie_pip > 0.5)'),options = list(pageLength = 10) )LD-mismatch
load(paste0("/project/xinhe/xsun/multi_group_ctwas/11.multi_group_1008/post_process_rm_ld/rmld_",trait,".rdata"))
sprintf("The number of problematic regions = %s", length(problematic_region_ids))[1] "The number of problematic regions = 2"sprintf("The number of problematic genes = %s", length(problematic_genes))[1] "The number of problematic genes = 2"sprintf("The number of problematic snps = %s", length(res_ldmismatch$problematic_snps))[1] "The number of problematic snps = 326"sprintf("The number of flipped snps = %s", length(res_ldmismatch$flipped_snps))[1] "The number of flipped snps = 3"finemap_rm_res_problematic_region <- finemap_res_rm[finemap_res_rm$id %in% problematic_genes,]
finemap_noLD_res_problematic_region <- res_rmld$finemap_res
merge_regionmerge_nold <- merge(finemap_rm_res_problematic_region,finemap_noLD_res_problematic_region, by = "id")
merge_regionmerge_nold <- merge_regionmerge_nold[,c("id","susie_pip.x","susie_pip.y")]
colnames(merge_regionmerge_nold) <- c("id","susie_pip_after_regionmerge","susie_pip_ld-mismatch-fixed")
DT::datatable(merge_regionmerge_nold,caption = htmltools::tags$caption(style = 'caption-side: topleft; text-align = left; color:black;','PIP after region merge and fixed PIP for problematic genes'),options = list(pageLength = 5) )finemap_res_origin <- ctwas_res_origin$finemap_res
finemap_res_origin_gene <- finemap_res_origin[finemap_res_origin$type != "SNP",]
p1 <- ggplot(data = finemap_res_origin_gene, aes(x= abs(z), y= susie_pip)) +
geom_point() +
ggtitle("Original ctwas results") +
theme_minimal()
finemap_res_rm_gene <- finemap_res_rm[finemap_res_rm$type != "SNP",]
p2 <- ggplot(data = finemap_res_rm_gene, aes(x= abs(z), y= susie_pip)) +
geom_point() +
ggtitle("After region merge") +
theme_minimal()
finemap_noLD_res_problematic_region_gene <- finemap_noLD_res_problematic_region[finemap_noLD_res_problematic_region$type !="SNP",]
p3 <- ggplot(data = finemap_noLD_res_problematic_region_gene, aes(x= abs(z), y= susie_pip)) +
geom_point() +
ggtitle("After LD mismatch fixed") +
theme_minimal()
grid.arrange(p1,p2,p3, ncol = 3)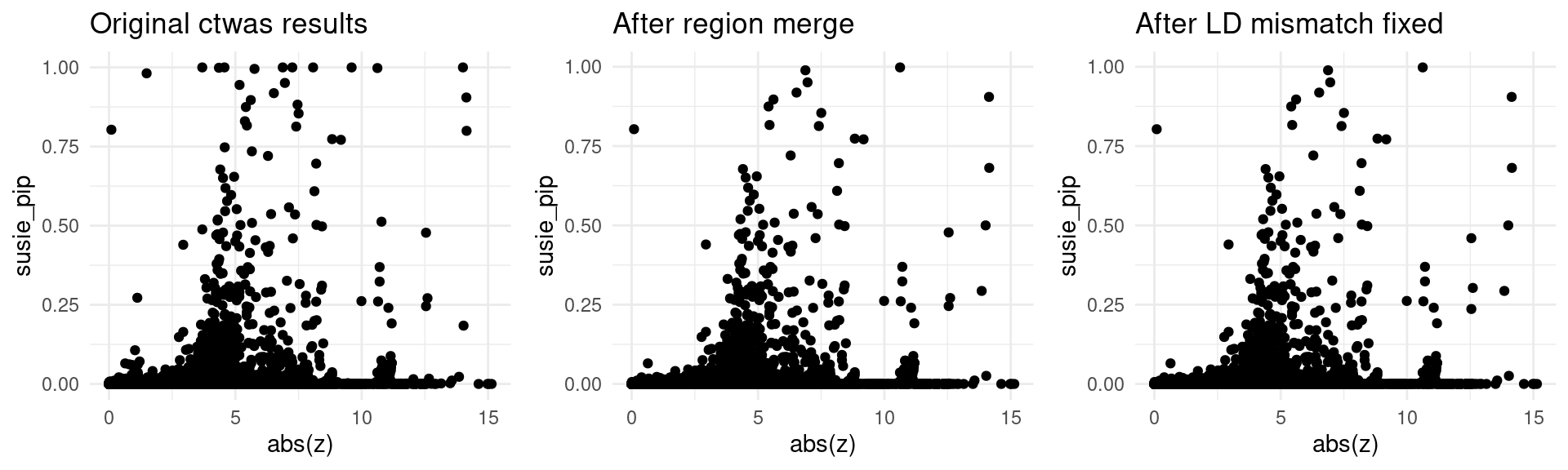
| Version | Author | Date |
|---|---|---|
| 4885dec | XSun | 2024-12-05 |
Comparing results without post processing
susie_alpha_res_origin <- ctwas_res_origin$susie_alpha_res
susie_alpha_res_origin <- anno_susie_alpha_res(susie_alpha_res_origin,
mapping_table = mapping_two,
map_by = "molecular_id",
drop_unmapped = TRUE)2024-12-06 13:23:46 INFO::Annotating susie alpha result ...
2024-12-06 13:23:46 INFO::Map molecular traits to genes
2024-12-06 13:23:46 INFO::Split PIPs for molecular traits mapped to multiple genescombined_pip_by_type_cs_origin <- combine_gene_pips(susie_alpha_res_origin,
group_by = "gene_name",
by = "type",
method = "combine_cs",
filter_cs = TRUE,
include_cs_id = T)
combined_pip_by_type_sig_origin <- combined_pip_by_type_cs_origin[combined_pip_by_type_cs_origin$combined_pip > 0.8,]
susie_alpha_res_rm <- res_regionmerge$susie_alpha_res
susie_alpha_res_rm <- anno_susie_alpha_res(susie_alpha_res_rm,
mapping_table = mapping_two,
map_by = "molecular_id",
drop_unmapped = TRUE)2024-12-06 13:23:49 INFO::Annotating susie alpha result ...
2024-12-06 13:23:49 INFO::Map molecular traits to genes
2024-12-06 13:23:49 INFO::Split PIPs for molecular traits mapped to multiple genescombined_pip_by_type_cs_rm <- combine_gene_pips(susie_alpha_res_rm,
group_by = "gene_name",
by = "type",
method = "combine_cs",
filter_cs = TRUE,
include_cs_id = T)
combined_pip_by_type_sig_rm <- combined_pip_by_type_cs_rm[combined_pip_by_type_cs_rm$combined_pip > 0.8,]
susie_alpha_res_rmld <- res_rmld$susie_alpha_res
susie_alpha_res_rmld <- anno_susie_alpha_res(susie_alpha_res_rmld,
mapping_table = mapping_two,
map_by = "molecular_id",
drop_unmapped = TRUE)2024-12-06 13:23:52 INFO::Annotating susie alpha result ...
2024-12-06 13:23:52 INFO::Map molecular traits to genes
2024-12-06 13:23:52 INFO::Split PIPs for molecular traits mapped to multiple genescombined_pip_by_type_cs_rmld <- combine_gene_pips(susie_alpha_res_rmld,
group_by = "gene_name",
by = "type",
method = "combine_cs",
filter_cs = TRUE,
include_cs_id = T)
combined_pip_by_type_sig_rmld <- combined_pip_by_type_cs_rmld[combined_pip_by_type_cs_rmld$combined_pip > 0.8,]
origin_unique <- combined_pip_by_type_sig_origin[!combined_pip_by_type_sig_origin$gene_name %in% combined_pip_by_type_sig_rmld$gene_name,]
origin_unique <- merge(origin_unique, combined_pip_by_type_cs_rm, by = "gene_name", all.x = TRUE)
# Then merge the result with combined_pip_by_type_cs_rmld
origin_unique <- merge(origin_unique, combined_pip_by_type_cs_rmld, by = "gene_name", all.x = TRUE)
origin_unique_show <- origin_unique[,c("gene_name","combined_pip.x","combined_pip.y","combined_pip","eQTL_pip.x","eQTL_pip.y","eQTL_pip","sQTL_pip.x","sQTL_pip.y","sQTL_pip","stQTL_pip.x","stQTL_pip.x","stQTL_pip","combined_cs_id.x","combined_cs_id.y","combined_cs_id")]
colnames(origin_unique_show) <- c("gene_name","combined_pip_origin","combined_pip_rm","combined_pip_rmld","eQTL_pip_origin","eQTL_pip_rm","eQTL_pip_rmld","sQTL_pip_origin","sQTL_pip_rm","sQTL_pip_rmld","stQTL_pip_origin","stQTL_pip_origin","stQTL_pip_rmld","combined_cs_id_origin","combined_cs_id_rm","combined_cs_id_rmld")
DT::datatable(origin_unique_show,caption = htmltools::tags$caption(style = 'caption-side: top; text-align: left; color: black;','Genes no longer significant after post processing'),options = list(pageLength = 5))Exploring allelic heterogeneity
pip_per_cs <- compute_pip_per_cs(combined_pip_by_type_sig_rmld, susie_alpha_res_rmld)
sprintf("Number of genes having allelic heterogeneity = %s",length(unique(pip_per_cs$gene_name)))[1] "Number of genes having allelic heterogeneity = 1"if(nrow(pip_per_cs) > 0) {
DT::datatable(pip_per_cs,caption = htmltools::tags$caption( style = 'caption-side: topleft; text-align = left; color:black;','PIP per CS'),options = list(pageLength = 5) )
}SBP-ukb-a-360
trait <- "SBP-ukb-a-360"
results_dir_origin <- paste0("/project/xinhe/xsun/multi_group_ctwas/11.multi_group_1008/results/",trait,"/")
ctwas_res_origin <- readRDS(paste0(results_dir_origin,trait,".ctwas.res.RDS"))
finemap_res_origin <- ctwas_res_origin$finemap_resRegion merge
load(paste0("/project/xinhe/xsun/multi_group_ctwas/11.multi_group_1008/post_process_rm_ld/rm_",trait,".rdata"))
finemap_res_rm <- res_regionmerge$finemap_res
finemap_res_rm_boundary_genes <- finemap_res_rm[finemap_res_rm$id %in%selected_boundary_genes$id,]
finemap_res_rm_boundary_genes_pip <- finemap_res_rm_boundary_genes[,c("id","susie_pip","cs")]
finemap_res_origin_boundary_genes <- finemap_res_origin[finemap_res_origin$id %in%selected_boundary_genes$id,]
finemap_res_origin_boundary_genes_pip <- finemap_res_origin_boundary_genes[,c("id","susie_pip","cs")]
finemap_res_compare_regionmerge <- merge(finemap_res_origin_boundary_genes_pip,finemap_res_rm_boundary_genes_pip, by = "id")
colnames(finemap_res_compare_regionmerge) <- c("id","susie_pip_origin","cs_origin","susie_pip_reginmerge","cs_reginmerge")
DT::datatable(finemap_res_compare_regionmerge,caption = htmltools::tags$caption( style = 'caption-side: left; text-align: left; color:black; font-size:150% ;','Selected boundary genes (susie_pip > 0.5)'),options = list(pageLength = 10) )LD-mismatch
load(paste0("/project/xinhe/xsun/multi_group_ctwas/11.multi_group_1008/post_process_rm_ld/rmld_",trait,".rdata"))
sprintf("The number of problematic regions = %s", length(problematic_region_ids))[1] "The number of problematic regions = 4"sprintf("The number of problematic genes = %s", length(problematic_genes))[1] "The number of problematic genes = 10"sprintf("The number of problematic snps = %s", length(res_ldmismatch$problematic_snps))[1] "The number of problematic snps = 742"sprintf("The number of flipped snps = %s", length(res_ldmismatch$flipped_snps))[1] "The number of flipped snps = 148"finemap_rm_res_problematic_region <- finemap_res_rm[finemap_res_rm$id %in% problematic_genes,]
finemap_noLD_res_problematic_region <- res_rmld$finemap_res
merge_regionmerge_nold <- merge(finemap_rm_res_problematic_region,finemap_noLD_res_problematic_region, by = "id")
merge_regionmerge_nold <- merge_regionmerge_nold[,c("id","susie_pip.x","susie_pip.y")]
colnames(merge_regionmerge_nold) <- c("id","susie_pip_after_regionmerge","susie_pip_ld-mismatch-fixed")
DT::datatable(merge_regionmerge_nold,caption = htmltools::tags$caption(style = 'caption-side: topleft; text-align = left; color:black;','PIP after region merge and fixed PIP for problematic genes'),options = list(pageLength = 5) )finemap_res_origin <- ctwas_res_origin$finemap_res
finemap_res_origin_gene <- finemap_res_origin[finemap_res_origin$type != "SNP",]
p1 <- ggplot(data = finemap_res_origin_gene, aes(x= abs(z), y= susie_pip)) +
geom_point() +
ggtitle("Original ctwas results") +
theme_minimal()
finemap_res_rm_gene <- finemap_res_rm[finemap_res_rm$type != "SNP",]
p2 <- ggplot(data = finemap_res_rm_gene, aes(x= abs(z), y= susie_pip)) +
geom_point() +
ggtitle("After region merge") +
theme_minimal()
finemap_noLD_res_problematic_region_gene <- finemap_noLD_res_problematic_region[finemap_noLD_res_problematic_region$type !="SNP",]
p3 <- ggplot(data = finemap_noLD_res_problematic_region_gene, aes(x= abs(z), y= susie_pip)) +
geom_point() +
ggtitle("After LD mismatch fixed") +
theme_minimal()
grid.arrange(p1,p2,p3, ncol = 3)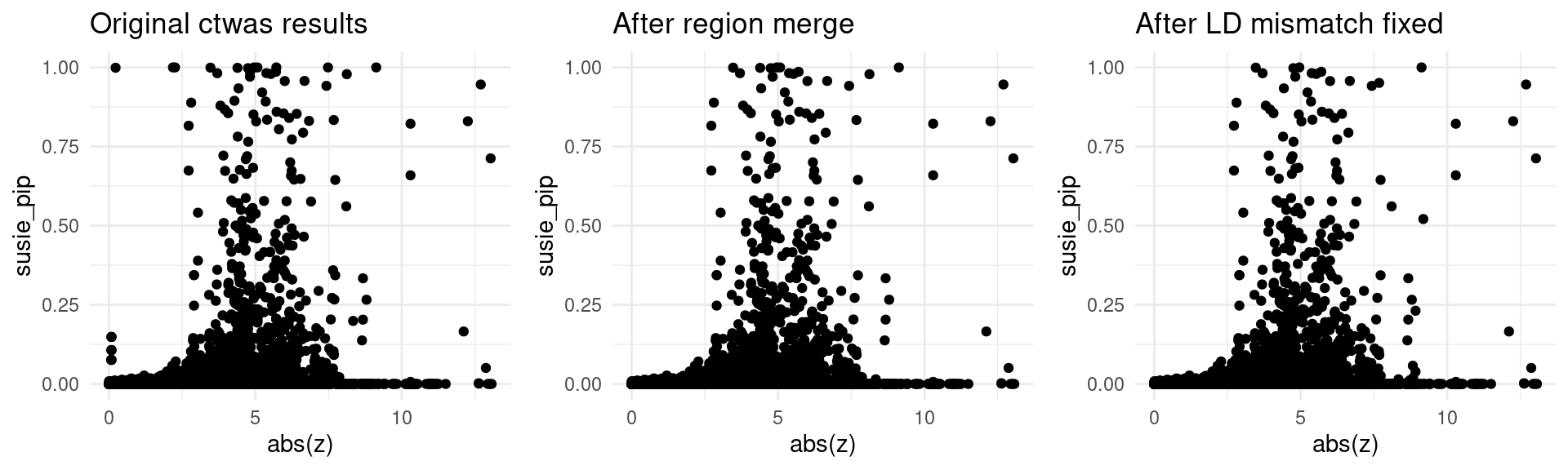
| Version | Author | Date |
|---|---|---|
| 4885dec | XSun | 2024-12-05 |
Comparing results without post processing
susie_alpha_res_origin <- ctwas_res_origin$susie_alpha_res
susie_alpha_res_origin <- anno_susie_alpha_res(susie_alpha_res_origin,
mapping_table = mapping_two,
map_by = "molecular_id",
drop_unmapped = TRUE)2024-12-06 13:24:42 INFO::Annotating susie alpha result ...
2024-12-06 13:24:42 INFO::Map molecular traits to genes
2024-12-06 13:24:43 INFO::Split PIPs for molecular traits mapped to multiple genescombined_pip_by_type_cs_origin <- combine_gene_pips(susie_alpha_res_origin,
group_by = "gene_name",
by = "type",
method = "combine_cs",
filter_cs = TRUE,
include_cs_id = T)
combined_pip_by_type_sig_origin <- combined_pip_by_type_cs_origin[combined_pip_by_type_cs_origin$combined_pip > 0.8,]
susie_alpha_res_rm <- res_regionmerge$susie_alpha_res
susie_alpha_res_rm <- anno_susie_alpha_res(susie_alpha_res_rm,
mapping_table = mapping_two,
map_by = "molecular_id",
drop_unmapped = TRUE)2024-12-06 13:24:49 INFO::Annotating susie alpha result ...
2024-12-06 13:24:49 INFO::Map molecular traits to genes
2024-12-06 13:24:50 INFO::Split PIPs for molecular traits mapped to multiple genescombined_pip_by_type_cs_rm <- combine_gene_pips(susie_alpha_res_rm,
group_by = "gene_name",
by = "type",
method = "combine_cs",
filter_cs = TRUE,
include_cs_id = T)
combined_pip_by_type_sig_rm <- combined_pip_by_type_cs_rm[combined_pip_by_type_cs_rm$combined_pip > 0.8,]
susie_alpha_res_rmld <- res_rmld$susie_alpha_res
susie_alpha_res_rmld <- anno_susie_alpha_res(susie_alpha_res_rmld,
mapping_table = mapping_two,
map_by = "molecular_id",
drop_unmapped = TRUE)2024-12-06 13:24:55 INFO::Annotating susie alpha result ...
2024-12-06 13:24:55 INFO::Map molecular traits to genes
2024-12-06 13:24:55 INFO::Split PIPs for molecular traits mapped to multiple genescombined_pip_by_type_cs_rmld <- combine_gene_pips(susie_alpha_res_rmld,
group_by = "gene_name",
by = "type",
method = "combine_cs",
filter_cs = TRUE,
include_cs_id = T)
combined_pip_by_type_sig_rmld <- combined_pip_by_type_cs_rmld[combined_pip_by_type_cs_rmld$combined_pip > 0.8,]
origin_unique <- combined_pip_by_type_sig_origin[!combined_pip_by_type_sig_origin$gene_name %in% combined_pip_by_type_sig_rmld$gene_name,]
origin_unique <- merge(origin_unique, combined_pip_by_type_cs_rm, by = "gene_name", all.x = TRUE)
# Then merge the result with combined_pip_by_type_cs_rmld
origin_unique <- merge(origin_unique, combined_pip_by_type_cs_rmld, by = "gene_name", all.x = TRUE)
origin_unique_show <- origin_unique[,c("gene_name","combined_pip.x","combined_pip.y","combined_pip","eQTL_pip.x","eQTL_pip.y","eQTL_pip","sQTL_pip.x","sQTL_pip.y","sQTL_pip","stQTL_pip.x","stQTL_pip.x","stQTL_pip","combined_cs_id.x","combined_cs_id.y","combined_cs_id")]
colnames(origin_unique_show) <- c("gene_name","combined_pip_origin","combined_pip_rm","combined_pip_rmld","eQTL_pip_origin","eQTL_pip_rm","eQTL_pip_rmld","sQTL_pip_origin","sQTL_pip_rm","sQTL_pip_rmld","stQTL_pip_origin","stQTL_pip_origin","stQTL_pip_rmld","combined_cs_id_origin","combined_cs_id_rm","combined_cs_id_rmld")
DT::datatable(origin_unique_show,caption = htmltools::tags$caption(style = 'caption-side: top; text-align: left; color: black;','Genes no longer significant after post processing'),options = list(pageLength = 5))Exploring allelic heterogeneity
pip_per_cs <- compute_pip_per_cs(combined_pip_by_type_sig_rmld, susie_alpha_res_rmld)
sprintf("Number of genes having allelic heterogeneity = %s",length(unique(pip_per_cs$gene_name)))[1] "Number of genes having allelic heterogeneity = 2"if(nrow(pip_per_cs) > 0) {
DT::datatable(pip_per_cs,caption = htmltools::tags$caption( style = 'caption-side: topleft; text-align = left; color:black;','PIP per CS'),options = list(pageLength = 5) )
}SCZ-ieu-b-5102
trait <- "SCZ-ieu-b-5102"
results_dir_origin <- paste0("/project/xinhe/xsun/multi_group_ctwas/11.multi_group_1008/results/",trait,"/")
ctwas_res_origin <- readRDS(paste0(results_dir_origin,trait,".ctwas.res.RDS"))
finemap_res_origin <- ctwas_res_origin$finemap_resRegion merge
load(paste0("/project/xinhe/xsun/multi_group_ctwas/11.multi_group_1008/post_process_rm_ld/rm_",trait,".rdata"))
finemap_res_rm <- res_regionmerge$finemap_res
finemap_res_rm_boundary_genes <- finemap_res_rm[finemap_res_rm$id %in%selected_boundary_genes$id,]
finemap_res_rm_boundary_genes_pip <- finemap_res_rm_boundary_genes[,c("id","susie_pip","cs")]
finemap_res_origin_boundary_genes <- finemap_res_origin[finemap_res_origin$id %in%selected_boundary_genes$id,]
finemap_res_origin_boundary_genes_pip <- finemap_res_origin_boundary_genes[,c("id","susie_pip","cs")]
finemap_res_compare_regionmerge <- merge(finemap_res_origin_boundary_genes_pip,finemap_res_rm_boundary_genes_pip, by = "id")
colnames(finemap_res_compare_regionmerge) <- c("id","susie_pip_origin","cs_origin","susie_pip_reginmerge","cs_reginmerge")
DT::datatable(finemap_res_compare_regionmerge,caption = htmltools::tags$caption( style = 'caption-side: left; text-align: left; color:black; font-size:150% ;','Selected boundary genes (susie_pip > 0.5)'),options = list(pageLength = 10) )LD-mismatch
load(paste0("/project/xinhe/xsun/multi_group_ctwas/11.multi_group_1008/post_process_rm_ld/rmld_",trait,".rdata"))
sprintf("The number of problematic regions = %s", length(problematic_region_ids))[1] "The number of problematic regions = 3"sprintf("The number of problematic genes = %s", length(problematic_genes))[1] "The number of problematic genes = 7"sprintf("The number of problematic snps = %s", length(res_ldmismatch$problematic_snps))[1] "The number of problematic snps = 169"sprintf("The number of flipped snps = %s", length(res_ldmismatch$flipped_snps))[1] "The number of flipped snps = 2"finemap_rm_res_problematic_region <- finemap_res_rm[finemap_res_rm$id %in% problematic_genes,]
finemap_noLD_res_problematic_region <- res_rmld$finemap_res
merge_regionmerge_nold <- merge(finemap_rm_res_problematic_region,finemap_noLD_res_problematic_region, by = "id")
merge_regionmerge_nold <- merge_regionmerge_nold[,c("id","susie_pip.x","susie_pip.y")]
colnames(merge_regionmerge_nold) <- c("id","susie_pip_after_regionmerge","susie_pip_ld-mismatch-fixed")
DT::datatable(merge_regionmerge_nold,caption = htmltools::tags$caption(style = 'caption-side: topleft; text-align = left; color:black;','PIP after region merge and fixed PIP for problematic genes'),options = list(pageLength = 5) )finemap_res_origin <- ctwas_res_origin$finemap_res
finemap_res_origin_gene <- finemap_res_origin[finemap_res_origin$type != "SNP",]
p1 <- ggplot(data = finemap_res_origin_gene, aes(x= abs(z), y= susie_pip)) +
geom_point() +
ggtitle("Original ctwas results") +
theme_minimal()
finemap_res_rm_gene <- finemap_res_rm[finemap_res_rm$type != "SNP",]
p2 <- ggplot(data = finemap_res_rm_gene, aes(x= abs(z), y= susie_pip)) +
geom_point() +
ggtitle("After region merge") +
theme_minimal()
finemap_noLD_res_problematic_region_gene <- finemap_noLD_res_problematic_region[finemap_noLD_res_problematic_region$type !="SNP",]
p3 <- ggplot(data = finemap_noLD_res_problematic_region_gene, aes(x= abs(z), y= susie_pip)) +
geom_point() +
ggtitle("After LD mismatch fixed") +
theme_minimal()
grid.arrange(p1,p2,p3, ncol = 3)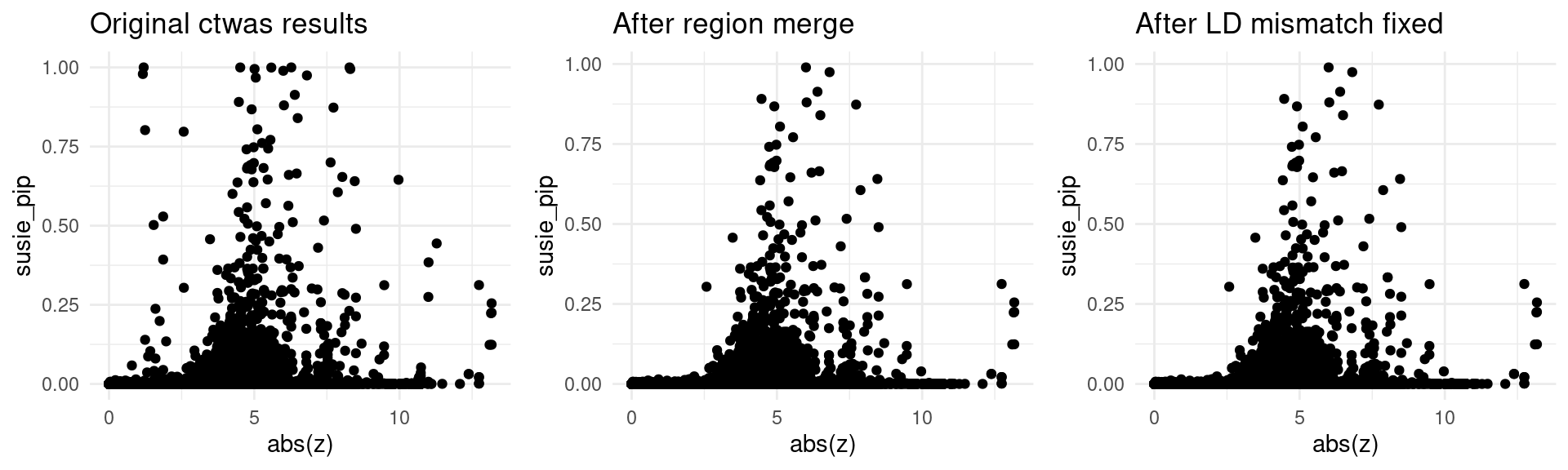
| Version | Author | Date |
|---|---|---|
| 4885dec | XSun | 2024-12-05 |
Comparing results without post processing
susie_alpha_res_origin <- ctwas_res_origin$susie_alpha_res
susie_alpha_res_origin <- anno_susie_alpha_res(susie_alpha_res_origin,
mapping_table = mapping_two,
map_by = "molecular_id",
drop_unmapped = TRUE)2024-12-06 13:25:42 INFO::Annotating susie alpha result ...
2024-12-06 13:25:42 INFO::Map molecular traits to genes
2024-12-06 13:25:42 INFO::Split PIPs for molecular traits mapped to multiple genescombined_pip_by_type_cs_origin <- combine_gene_pips(susie_alpha_res_origin,
group_by = "gene_name",
by = "type",
method = "combine_cs",
filter_cs = TRUE,
include_cs_id = T)
combined_pip_by_type_sig_origin <- combined_pip_by_type_cs_origin[combined_pip_by_type_cs_origin$combined_pip > 0.8,]
susie_alpha_res_rm <- res_regionmerge$susie_alpha_res
susie_alpha_res_rm <- anno_susie_alpha_res(susie_alpha_res_rm,
mapping_table = mapping_two,
map_by = "molecular_id",
drop_unmapped = TRUE)2024-12-06 13:25:45 INFO::Annotating susie alpha result ...
2024-12-06 13:25:45 INFO::Map molecular traits to genes
2024-12-06 13:25:46 INFO::Split PIPs for molecular traits mapped to multiple genescombined_pip_by_type_cs_rm <- combine_gene_pips(susie_alpha_res_rm,
group_by = "gene_name",
by = "type",
method = "combine_cs",
filter_cs = TRUE,
include_cs_id = T)
combined_pip_by_type_sig_rm <- combined_pip_by_type_cs_rm[combined_pip_by_type_cs_rm$combined_pip > 0.8,]
susie_alpha_res_rmld <- res_rmld$susie_alpha_res
susie_alpha_res_rmld <- anno_susie_alpha_res(susie_alpha_res_rmld,
mapping_table = mapping_two,
map_by = "molecular_id",
drop_unmapped = TRUE)2024-12-06 13:25:48 INFO::Annotating susie alpha result ...
2024-12-06 13:25:48 INFO::Map molecular traits to genes
2024-12-06 13:25:49 INFO::Split PIPs for molecular traits mapped to multiple genescombined_pip_by_type_cs_rmld <- combine_gene_pips(susie_alpha_res_rmld,
group_by = "gene_name",
by = "type",
method = "combine_cs",
filter_cs = TRUE,
include_cs_id = T)
combined_pip_by_type_sig_rmld <- combined_pip_by_type_cs_rmld[combined_pip_by_type_cs_rmld$combined_pip > 0.8,]
origin_unique <- combined_pip_by_type_sig_origin[!combined_pip_by_type_sig_origin$gene_name %in% combined_pip_by_type_sig_rmld$gene_name,]
origin_unique <- merge(origin_unique, combined_pip_by_type_cs_rm, by = "gene_name", all.x = TRUE)
# Then merge the result with combined_pip_by_type_cs_rmld
origin_unique <- merge(origin_unique, combined_pip_by_type_cs_rmld, by = "gene_name", all.x = TRUE)
origin_unique_show <- origin_unique[,c("gene_name","combined_pip.x","combined_pip.y","combined_pip","eQTL_pip.x","eQTL_pip.y","eQTL_pip","sQTL_pip.x","sQTL_pip.y","sQTL_pip","stQTL_pip.x","stQTL_pip.x","stQTL_pip","combined_cs_id.x","combined_cs_id.y","combined_cs_id")]
colnames(origin_unique_show) <- c("gene_name","combined_pip_origin","combined_pip_rm","combined_pip_rmld","eQTL_pip_origin","eQTL_pip_rm","eQTL_pip_rmld","sQTL_pip_origin","sQTL_pip_rm","sQTL_pip_rmld","stQTL_pip_origin","stQTL_pip_origin","stQTL_pip_rmld","combined_cs_id_origin","combined_cs_id_rm","combined_cs_id_rmld")
DT::datatable(origin_unique_show,caption = htmltools::tags$caption(style = 'caption-side: top; text-align: left; color: black;','Genes no longer significant after post processing'),options = list(pageLength = 5))Exploring allelic heterogeneity
pip_per_cs <- compute_pip_per_cs(combined_pip_by_type_sig_rmld, susie_alpha_res_rmld)
sprintf("Number of genes having allelic heterogeneity = %s",length(unique(pip_per_cs$gene_name)))[1] "Number of genes having allelic heterogeneity = 0"if(nrow(pip_per_cs) > 0) {
DT::datatable(pip_per_cs,caption = htmltools::tags$caption( style = 'caption-side: topleft; text-align = left; color:black;','PIP per CS'),options = list(pageLength = 5) )
}WBC-ieu-b-30
trait <- "WBC-ieu-b-30"
results_dir_origin <- paste0("/project/xinhe/xsun/multi_group_ctwas/11.multi_group_1008/results/",trait,"/")
ctwas_res_origin <- readRDS(paste0(results_dir_origin,trait,".ctwas.res.RDS"))
finemap_res_origin <- ctwas_res_origin$finemap_resRegion merge
load(paste0("/project/xinhe/xsun/multi_group_ctwas/11.multi_group_1008/post_process_rm_ld/rm_",trait,".rdata"))
finemap_res_rm <- res_regionmerge$finemap_res
finemap_res_rm_boundary_genes <- finemap_res_rm[finemap_res_rm$id %in%selected_boundary_genes$id,]
finemap_res_rm_boundary_genes_pip <- finemap_res_rm_boundary_genes[,c("id","susie_pip","cs")]
finemap_res_origin_boundary_genes <- finemap_res_origin[finemap_res_origin$id %in%selected_boundary_genes$id,]
finemap_res_origin_boundary_genes_pip <- finemap_res_origin_boundary_genes[,c("id","susie_pip","cs")]
finemap_res_compare_regionmerge <- merge(finemap_res_origin_boundary_genes_pip,finemap_res_rm_boundary_genes_pip, by = "id")
colnames(finemap_res_compare_regionmerge) <- c("id","susie_pip_origin","cs_origin","susie_pip_reginmerge","cs_reginmerge")
DT::datatable(finemap_res_compare_regionmerge,caption = htmltools::tags$caption( style = 'caption-side: left; text-align: left; color:black; font-size:150% ;','Selected boundary genes (susie_pip > 0.5)'),options = list(pageLength = 10) )LD-mismatch
load(paste0("/project/xinhe/xsun/multi_group_ctwas/11.multi_group_1008/post_process_rm_ld/rmld_",trait,".rdata"))
sprintf("The number of problematic regions = %s", length(problematic_region_ids))[1] "The number of problematic regions = 16"sprintf("The number of problematic genes = %s", length(problematic_genes))[1] "The number of problematic genes = 37"sprintf("The number of problematic snps = %s", length(res_ldmismatch$problematic_snps))[1] "The number of problematic snps = 1092"sprintf("The number of flipped snps = %s", length(res_ldmismatch$flipped_snps))[1] "The number of flipped snps = 17"finemap_rm_res_problematic_region <- finemap_res_rm[finemap_res_rm$id %in% problematic_genes,]
finemap_noLD_res_problematic_region <- res_rmld$finemap_res
merge_regionmerge_nold <- merge(finemap_rm_res_problematic_region,finemap_noLD_res_problematic_region, by = "id")
merge_regionmerge_nold <- merge_regionmerge_nold[,c("id","susie_pip.x","susie_pip.y")]
colnames(merge_regionmerge_nold) <- c("id","susie_pip_after_regionmerge","susie_pip_ld-mismatch-fixed")
DT::datatable(merge_regionmerge_nold,caption = htmltools::tags$caption(style = 'caption-side: topleft; text-align = left; color:black;','PIP after region merge and fixed PIP for problematic genes'),options = list(pageLength = 5) )finemap_res_origin <- ctwas_res_origin$finemap_res
finemap_res_origin_gene <- finemap_res_origin[finemap_res_origin$type != "SNP",]
p1 <- ggplot(data = finemap_res_origin_gene, aes(x= abs(z), y= susie_pip)) +
geom_point() +
ggtitle("Original ctwas results") +
theme_minimal()
finemap_res_rm_gene <- finemap_res_rm[finemap_res_rm$type != "SNP",]
p2 <- ggplot(data = finemap_res_rm_gene, aes(x= abs(z), y= susie_pip)) +
geom_point() +
ggtitle("After region merge") +
theme_minimal()
finemap_noLD_res_problematic_region_gene <- finemap_noLD_res_problematic_region[finemap_noLD_res_problematic_region$type !="SNP",]
p3 <- ggplot(data = finemap_noLD_res_problematic_region_gene, aes(x= abs(z), y= susie_pip)) +
geom_point() +
ggtitle("After LD mismatch fixed") +
theme_minimal()
grid.arrange(p1,p2,p3, ncol = 3)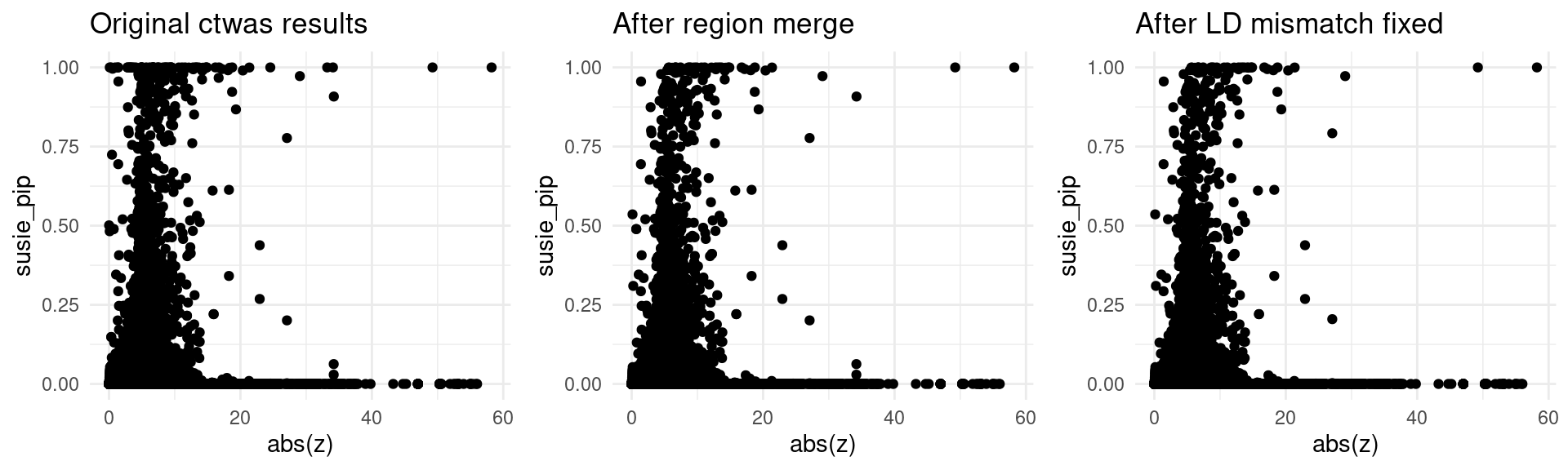
| Version | Author | Date |
|---|---|---|
| 984b87f | XSun | 2024-12-06 |
Comparing results without post processing
susie_alpha_res_origin <- ctwas_res_origin$susie_alpha_res
susie_alpha_res_origin <- anno_susie_alpha_res(susie_alpha_res_origin,
mapping_table = mapping_two,
map_by = "molecular_id",
drop_unmapped = TRUE)2024-12-06 13:27:08 INFO::Annotating susie alpha result ...
2024-12-06 13:27:08 INFO::Map molecular traits to genes
2024-12-06 13:27:09 INFO::Split PIPs for molecular traits mapped to multiple genescombined_pip_by_type_cs_origin <- combine_gene_pips(susie_alpha_res_origin,
group_by = "gene_name",
by = "type",
method = "combine_cs",
filter_cs = TRUE,
include_cs_id = T)
combined_pip_by_type_sig_origin <- combined_pip_by_type_cs_origin[combined_pip_by_type_cs_origin$combined_pip > 0.8,]
susie_alpha_res_rm <- res_regionmerge$susie_alpha_res
susie_alpha_res_rm <- anno_susie_alpha_res(susie_alpha_res_rm,
mapping_table = mapping_two,
map_by = "molecular_id",
drop_unmapped = TRUE)2024-12-06 13:27:21 INFO::Annotating susie alpha result ...
2024-12-06 13:27:21 INFO::Map molecular traits to genes
2024-12-06 13:27:22 INFO::Split PIPs for molecular traits mapped to multiple genescombined_pip_by_type_cs_rm <- combine_gene_pips(susie_alpha_res_rm,
group_by = "gene_name",
by = "type",
method = "combine_cs",
filter_cs = TRUE,
include_cs_id = T)
combined_pip_by_type_sig_rm <- combined_pip_by_type_cs_rm[combined_pip_by_type_cs_rm$combined_pip > 0.8,]
susie_alpha_res_rmld <- res_rmld$susie_alpha_res
susie_alpha_res_rmld <- anno_susie_alpha_res(susie_alpha_res_rmld,
mapping_table = mapping_two,
map_by = "molecular_id",
drop_unmapped = TRUE)2024-12-06 13:27:34 INFO::Annotating susie alpha result ...
2024-12-06 13:27:34 INFO::Map molecular traits to genes
2024-12-06 13:27:35 INFO::Split PIPs for molecular traits mapped to multiple genescombined_pip_by_type_cs_rmld <- combine_gene_pips(susie_alpha_res_rmld,
group_by = "gene_name",
by = "type",
method = "combine_cs",
filter_cs = TRUE,
include_cs_id = T)
combined_pip_by_type_sig_rmld <- combined_pip_by_type_cs_rmld[combined_pip_by_type_cs_rmld$combined_pip > 0.8,]
origin_unique <- combined_pip_by_type_sig_origin[!combined_pip_by_type_sig_origin$gene_name %in% combined_pip_by_type_sig_rmld$gene_name,]
origin_unique <- merge(origin_unique, combined_pip_by_type_cs_rm, by = "gene_name", all.x = TRUE)
# Then merge the result with combined_pip_by_type_cs_rmld
origin_unique <- merge(origin_unique, combined_pip_by_type_cs_rmld, by = "gene_name", all.x = TRUE)
origin_unique_show <- origin_unique[,c("gene_name","combined_pip.x","combined_pip.y","combined_pip","eQTL_pip.x","eQTL_pip.y","eQTL_pip","sQTL_pip.x","sQTL_pip.y","sQTL_pip","stQTL_pip.x","stQTL_pip.x","stQTL_pip","combined_cs_id.x","combined_cs_id.y","combined_cs_id")]
colnames(origin_unique_show) <- c("gene_name","combined_pip_origin","combined_pip_rm","combined_pip_rmld","eQTL_pip_origin","eQTL_pip_rm","eQTL_pip_rmld","sQTL_pip_origin","sQTL_pip_rm","sQTL_pip_rmld","stQTL_pip_origin","stQTL_pip_origin","stQTL_pip_rmld","combined_cs_id_origin","combined_cs_id_rm","combined_cs_id_rmld")
DT::datatable(origin_unique_show,caption = htmltools::tags$caption(style = 'caption-side: top; text-align: left; color: black;','Genes no longer significant after post processing'),options = list(pageLength = 5))Exploring allelic heterogeneity
pip_per_cs <- compute_pip_per_cs(combined_pip_by_type_sig_rmld, susie_alpha_res_rmld)
sprintf("Number of genes having allelic heterogeneity = %s",length(unique(pip_per_cs$gene_name)))[1] "Number of genes having allelic heterogeneity = 12"if(nrow(pip_per_cs) > 0) {
DT::datatable(pip_per_cs,caption = htmltools::tags$caption( style = 'caption-side: topleft; text-align = left; color:black;','PIP per CS'),options = list(pageLength = 5) )
}
sessionInfo()R version 4.2.0 (2022-04-22)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] C
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] dplyr_1.1.4 gridExtra_2.3
[3] ggplot2_3.5.1 EnsDb.Hsapiens.v86_2.99.0
[5] ensembldb_2.20.2 AnnotationFilter_1.20.0
[7] GenomicFeatures_1.48.3 AnnotationDbi_1.58.0
[9] Biobase_2.56.0 GenomicRanges_1.48.0
[11] GenomeInfoDb_1.39.9 IRanges_2.30.0
[13] S4Vectors_0.34.0 BiocGenerics_0.42.0
[15] ctwas_0.4.20
loaded via a namespace (and not attached):
[1] colorspace_2.0-3 rjson_0.2.21
[3] ellipsis_0.3.2 rprojroot_2.0.3
[5] XVector_0.36.0 locuszoomr_0.2.1
[7] fs_1.5.2 rstudioapi_0.13
[9] farver_2.1.0 DT_0.22
[11] ggrepel_0.9.1 bit64_4.0.5
[13] fansi_1.0.3 xml2_1.3.3
[15] codetools_0.2-18 logging_0.10-108
[17] cachem_1.0.6 knitr_1.39
[19] jsonlite_1.8.0 workflowr_1.7.0
[21] Rsamtools_2.12.0 dbplyr_2.1.1
[23] png_0.1-7 readr_2.1.2
[25] compiler_4.2.0 httr_1.4.3
[27] assertthat_0.2.1 Matrix_1.5-3
[29] fastmap_1.1.0 lazyeval_0.2.2
[31] cli_3.6.1 later_1.3.0
[33] htmltools_0.5.2 prettyunits_1.1.1
[35] tools_4.2.0 gtable_0.3.0
[37] glue_1.6.2 GenomeInfoDbData_1.2.8
[39] rappdirs_0.3.3 Rcpp_1.0.12
[41] jquerylib_0.1.4 vctrs_0.6.5
[43] Biostrings_2.64.0 rtracklayer_1.56.0
[45] crosstalk_1.2.0 xfun_0.41
[47] stringr_1.5.1 lifecycle_1.0.4
[49] irlba_2.3.5 restfulr_0.0.14
[51] XML_3.99-0.14 zlibbioc_1.42.0
[53] zoo_1.8-10 scales_1.3.0
[55] gggrid_0.2-0 hms_1.1.1
[57] promises_1.2.0.1 MatrixGenerics_1.8.0
[59] ProtGenerics_1.28.0 parallel_4.2.0
[61] SummarizedExperiment_1.26.1 LDlinkR_1.2.3
[63] yaml_2.3.5 curl_4.3.2
[65] memoise_2.0.1 sass_0.4.1
[67] biomaRt_2.54.1 stringi_1.7.6
[69] RSQLite_2.3.1 highr_0.9
[71] BiocIO_1.6.0 filelock_1.0.2
[73] BiocParallel_1.30.3 rlang_1.1.2
[75] pkgconfig_2.0.3 matrixStats_0.62.0
[77] bitops_1.0-7 evaluate_0.15
[79] lattice_0.20-45 purrr_1.0.2
[81] labeling_0.4.2 GenomicAlignments_1.32.0
[83] htmlwidgets_1.5.4 cowplot_1.1.1
[85] bit_4.0.4 tidyselect_1.2.0
[87] magrittr_2.0.3 R6_2.5.1
[89] generics_0.1.2 DelayedArray_0.22.0
[91] DBI_1.2.2 withr_2.5.0
[93] pgenlibr_0.3.3 pillar_1.9.0
[95] whisker_0.4 KEGGREST_1.36.3
[97] RCurl_1.98-1.7 mixsqp_0.3-43
[99] tibble_3.2.1 crayon_1.5.1
[101] utf8_1.2.2 BiocFileCache_2.4.0
[103] plotly_4.10.0 tzdb_0.4.0
[105] rmarkdown_2.25 progress_1.2.2
[107] grid_4.2.0 data.table_1.14.2
[109] blob_1.2.3 git2r_0.30.1
[111] digest_0.6.29 tidyr_1.3.0
[113] httpuv_1.6.5 munsell_0.5.0
[115] viridisLite_0.4.0 bslib_0.3.1