Shared effect size parameters
30% PVE and 2.5e-4 prior inclusion for SNPs, 5% PVE and 0.015 prior inclusion for Liver, 1% PVE and 0.003 prior inclusion for other three mixed groups.
results_dir <- "/project/xinhe/shengqian/cTWAS_simulation/simulation_same_two_tissues/"
runtag = "ukb-s80.45-3_corr"
configtag <- 1
simutags <- paste(1, 1:5, sep = "-")
thin <- 0.1
sample_size <- 45000
PIP_threshold <- 0.8#results using PIP threshold (gene+tissue)
results_df[,c("simutag", "n_causal", "n_detected_pip", "n_detected_pip_in_causal")] simutag n_causal n_detected_pip n_detected_pip_in_causal
1 1-1 118 0 0
2 1-2 95 0 0
3 1-3 116 0 0
4 1-4 117 0 0
5 1-5 113 0 0#mean percent causal using PIP > 0.8
sum(results_df$n_detected_pip_in_causal)/sum(results_df$n_detected_pip)[1] NaN#results using combined PIP threshold
results_df[,c("simutag", "n_causal_combined", "n_detected_comb_pip", "n_detected_comb_pip_in_causal")] simutag n_causal_combined n_detected_comb_pip n_detected_comb_pip_in_causal
1 1-1 118 31 28
2 1-2 95 32 28
3 1-3 116 26 26
4 1-4 117 41 36
5 1-5 113 40 33#mean percent causal using combined PIP > 0.8
sum(results_df$n_detected_comb_pip_in_causal)/sum(results_df$n_detected_comb_pip)[1] 0.8882353#prior inclusion and mean prior inclusion
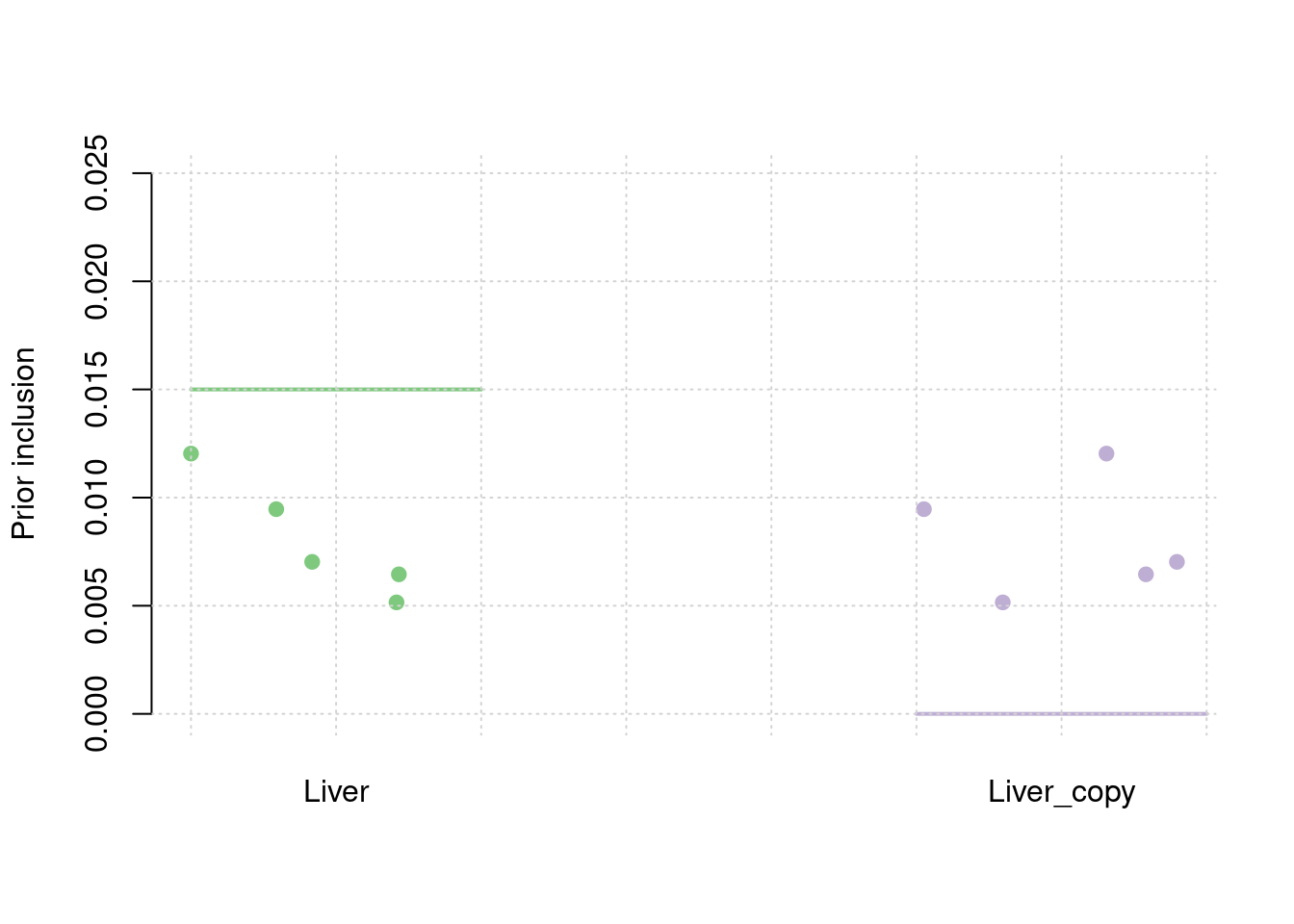
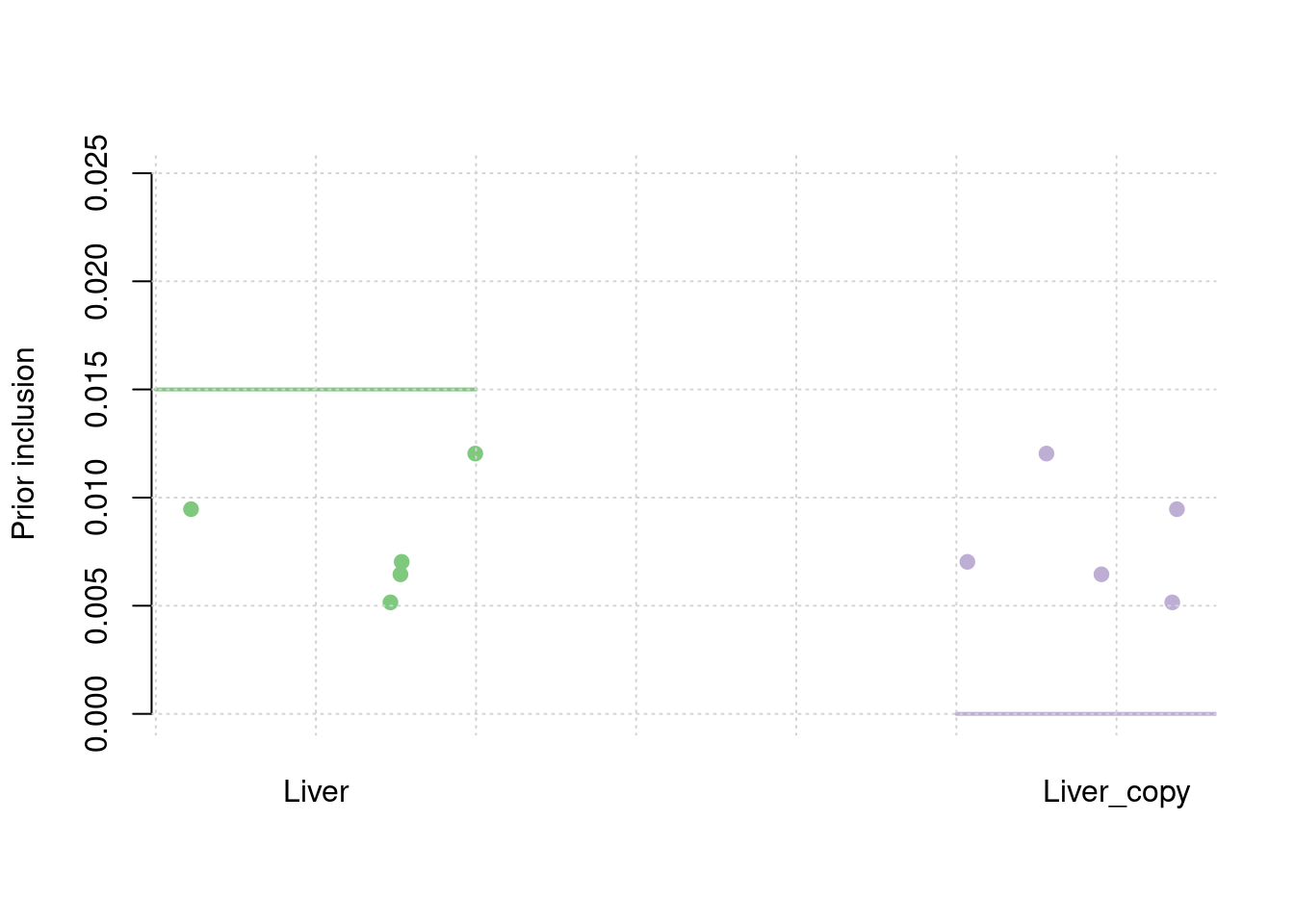
results_df[,c(which(colnames(results_df)=="simutag"), setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df))))] simutag prior_snp prior_weight1 prior_weight2
1 1-1 0.0002108796 0.005154730 0.005154730
2 1-2 0.0002573964 0.007027990 0.007027990
3 1-3 0.0002761674 0.006451763 0.006451763
4 1-4 0.0002896721 0.012034873 0.012034873
5 1-5 0.0002807151 0.009465010 0.009465010colMeans(results_df[,setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df)))]) prior_snp prior_weight1 prior_weight2
0.0002629661 0.0080268734 0.0080268734 #prior variance and mean prior variance
results_df[,c(which(colnames(results_df)=="simutag"), grep("prior_var", names(results_df)))] simutag prior_var_snp prior_var_weight1 prior_var_weight2
1 1-1 9.443700 32.95046 32.95046
2 1-2 8.605591 22.28596 22.28596
3 1-3 7.982523 26.83210 26.83210
4 1-4 7.486181 14.84881 14.84881
5 1-5 7.210972 21.07834 21.07834colMeans(results_df[,grep("prior_var", names(results_df))]) prior_var_snp prior_var_weight1 prior_var_weight2
8.145793 23.599134 23.599134 #PVE and mean PVE
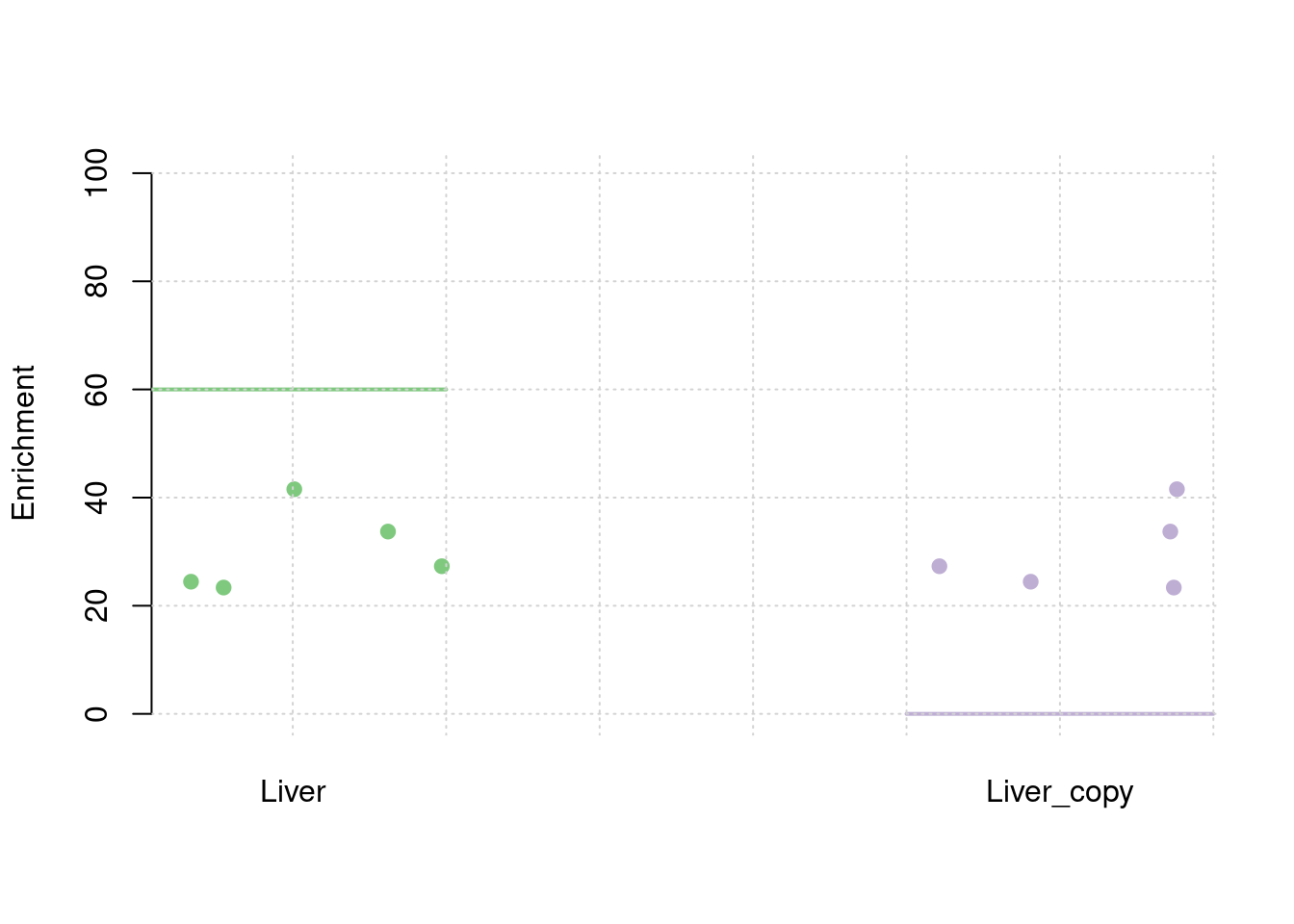
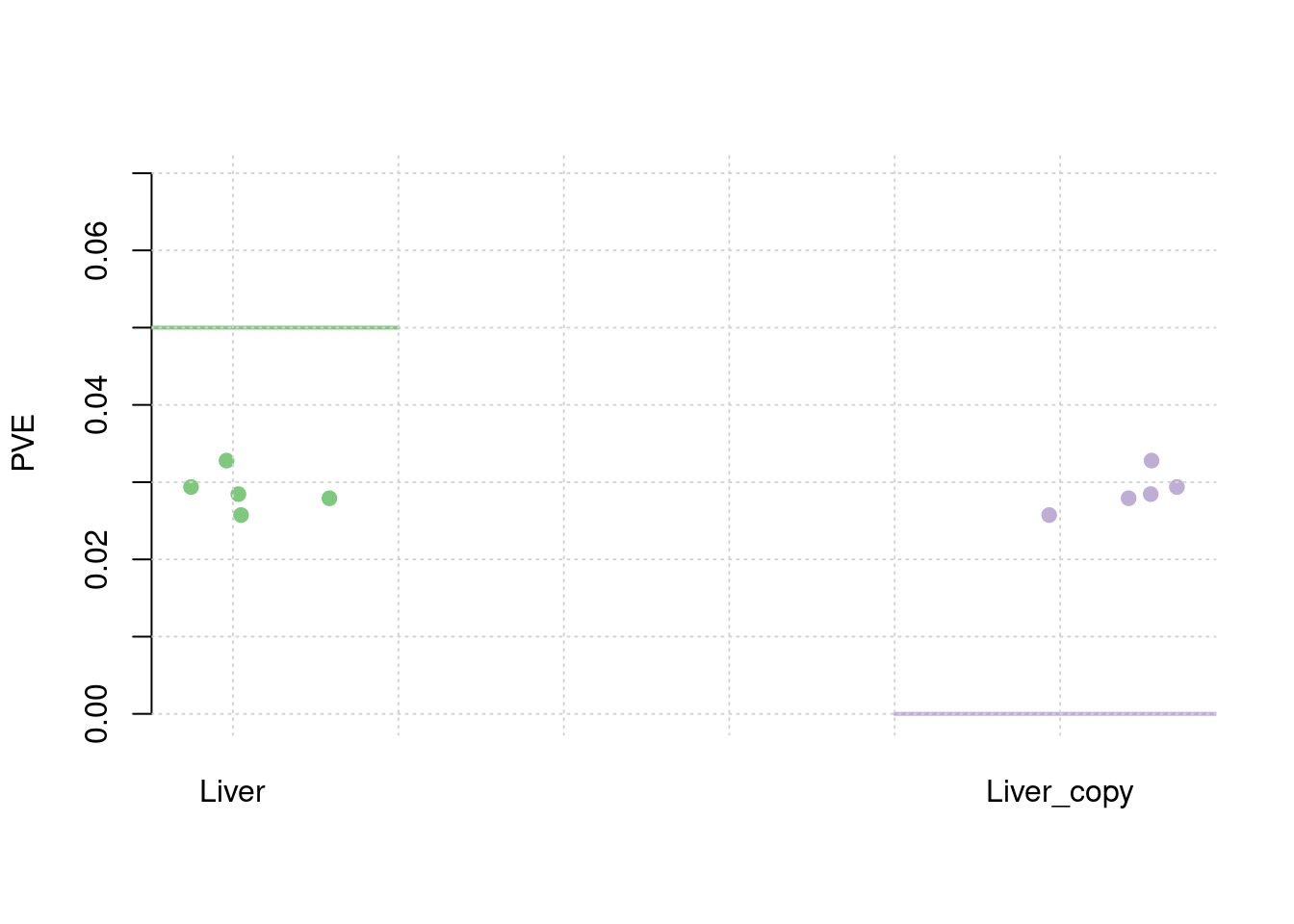
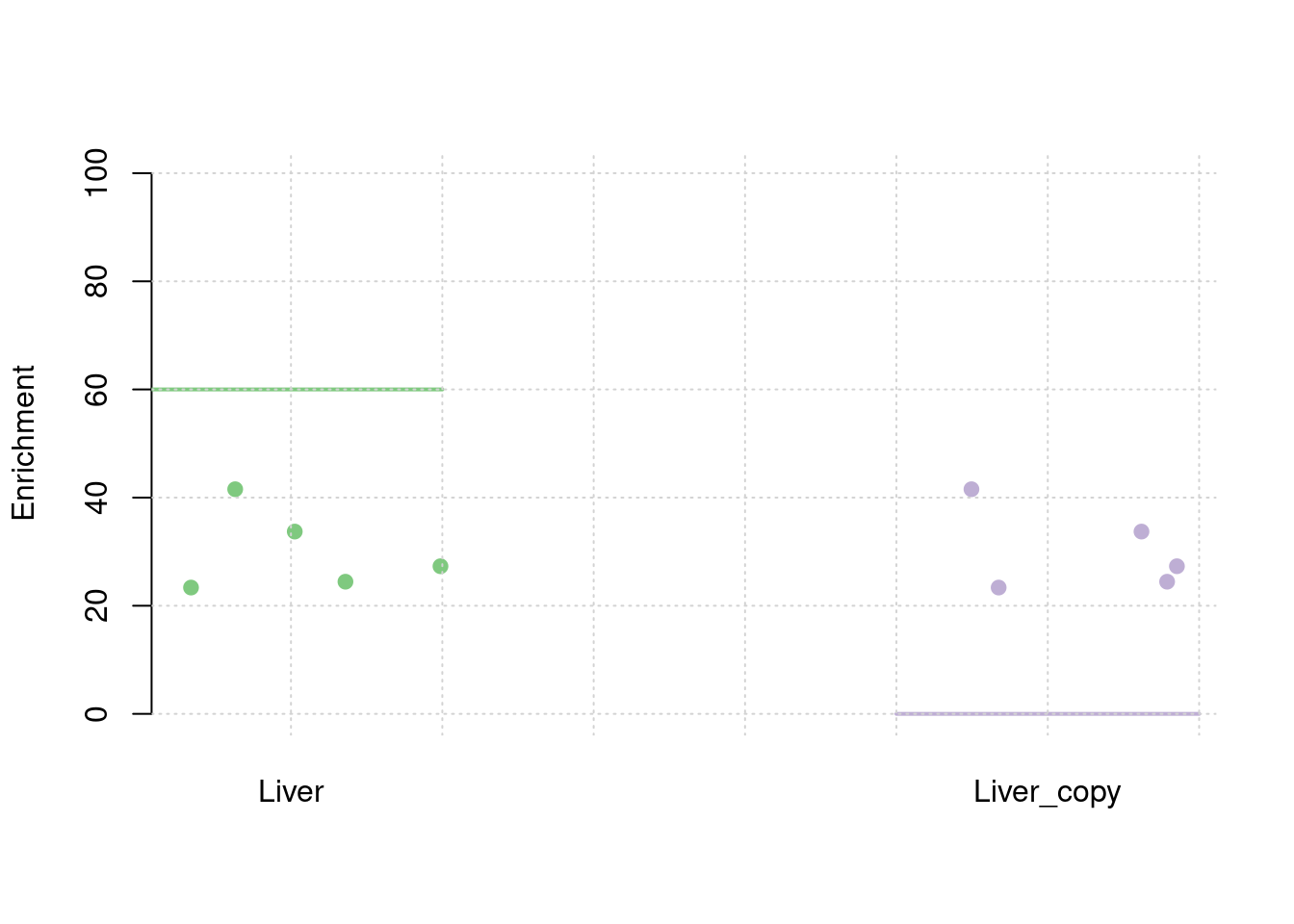
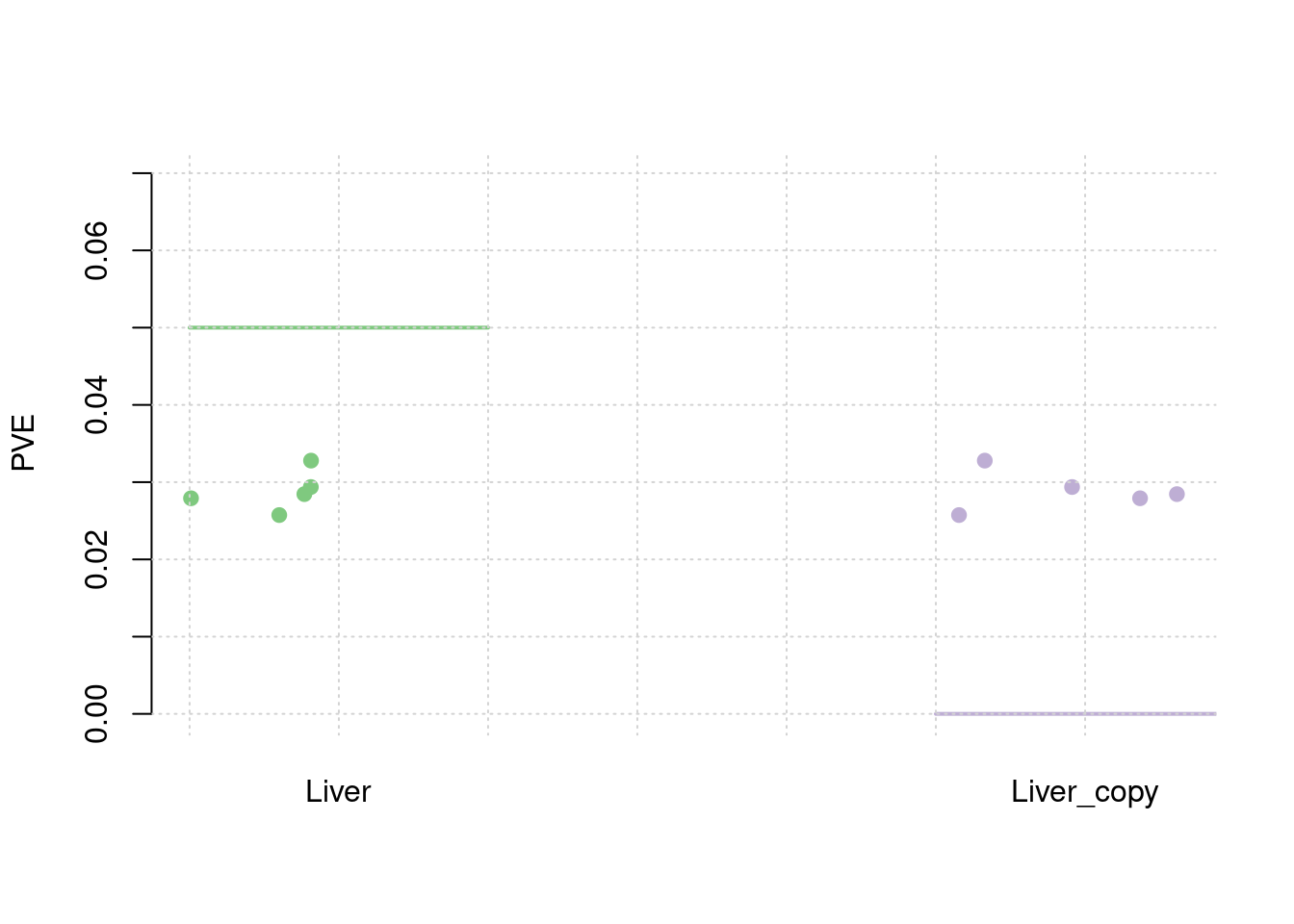
results_df[,c(which(colnames(results_df)=="simutag"), grep("pve", names(results_df)))] simutag pve_snp pve_weight1 pve_weight2
1 1-1 0.2344277 0.02791214 0.02791214
2 1-2 0.2607447 0.02573879 0.02573879
3 1-3 0.2595045 0.02844846 0.02844846
4 1-4 0.2552697 0.02936695 0.02936695
5 1-5 0.2382823 0.03278560 0.03278560colMeans(results_df[,grep("pve", names(results_df))]) pve_snp pve_weight1 pve_weight2
0.24964577 0.02885039 0.02885039 #TWAS results
results_df[,c(which(colnames(results_df)=="simutag"), grep("twas", names(results_df)))] simutag n_detected_twas n_detected_twas_in_causal n_detected_comb_twas
1 1-1 136 34 68
2 1-2 146 34 73
3 1-3 150 37 75
4 1-4 168 44 84
5 1-5 146 38 73
n_detected_comb_twas_in_causal
1 34
2 34
3 37
4 44
5 38sum(results_df$n_detected_comb_twas_in_causal)/sum(results_df$n_detected_comb_twas)[1] 0.5013405
Separate effect size parameters
For the cTWAS analysis, each tissue had its own prior inclusion parameter end effect size parameter.
results_dir <- "/project/xinhe/shengqian/cTWAS_simulation/simulation_same_two_tissues/"
runtag = "ukb-s80.45-3_corr"
configtag <- 2
simutags <- paste(1, 1:5, sep = "-")
thin <- 0.1
sample_size <- 45000
PIP_threshold <- 0.8results_df <- data.frame(simutag=as.character(),
n_causal=as.integer(),
n_causal_combined=as.integer(),
n_detected_pip=as.integer(),
n_detected_pip_in_causal=as.integer(),
n_detected_comb_pip=as.integer(),
n_detected_comb_pip_in_causal=as.integer(),
pve_snp=as.numeric(),
pve_weight1=as.numeric(),
pve_weight2=as.numeric(),
prior_weight1=as.numeric(),
prior_weight2=as.numeric(),
prior_var_snp=as.numeric(),
prior_var_weight1=as.numeric(),
prior_var_weight2=as.numeric(),
n_detected_twas=as.integer(),
n_detected_twas_in_causal=as.integer(),
n_detected_comb_twas=as.integer(),
n_detected_comb_twas_in_causal=as.integer())
for (i in 1:length(simutags)){
simutag <- simutags[i]
#load genes with true simulated effect
load(paste0(results_dir, runtag, "_simu", simutag, "-pheno.Rd"))
true_genes <- unlist(sapply(1:22, function(x){phenores$batch[[x]]$id.cgene}))
true_genes_combined <- unique(sapply(true_genes, function(x){unlist(strsplit(x, "[|]"))[1]}))
#load cTWAS results
ctwas_res <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR.susieIrss.txt"))
ctwas_gene_res <- ctwas_res[ctwas_res$type!="SNP",]
#number of causal genes
n_causal <- length(true_genes)
n_causal_combined <- length(true_genes_combined)
#number of gene+tissue combinations with cTWAS PIP > threshold
n_ctwas_genes <- sum(ctwas_gene_res$susie_pip > PIP_threshold)
#number of cTWAS genes that are causal
n_causal_detected <- sum(ctwas_gene_res$id[ctwas_gene_res$susie_pip > PIP_threshold] %in% true_genes)
#collapse gene+tissues to genes and compute combined PIP
ctwas_gene_res$gene <- sapply(ctwas_gene_res$id, function(x){unlist(strsplit(x,"[|]"))[1]})
ctwas_gene_res_combined <- aggregate(ctwas_gene_res$susie_pip, by=list(ctwas_gene_res$gene), FUN=sum)
colnames(ctwas_gene_res_combined) <- c("gene", "pip_combined")
#number of genes with combined PIP > threshold
n_ctwas_genes_combined <- sum(ctwas_gene_res_combined$pip_combined > PIP_threshold)
#number of cTWAS genes using combined PIP that are causal
n_causal_detected_combined <- sum(ctwas_gene_res_combined$gene[ctwas_gene_res_combined$pip_combined > PIP_threshold] %in% true_genes_combined)
#collect number of SNPs analyzed by cTWAS
ctwas_res_s1 <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR.s1.susieIrss.txt"))
n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
rm(ctwas_res_s1)
#load estimated parameters
load(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR.s2.susieIrssres.Rd"))
#estimated group prior (all iterations)
estimated_group_prior_all <- group_prior_rec
estimated_group_prior_all["SNP",] <- estimated_group_prior_all["SNP",]*thin #adjust parameter to account for thin argument
#estimated group prior variance (all iterations)
estimated_group_prior_var_all <- group_prior_var_rec
#set group size
group_size <- c(table(ctwas_gene_res$type), structure(n_snps, names="SNP"))
group_size <- group_size[rownames(estimated_group_prior_all)]
#estimated group PVE (all iterations)
estimated_group_pve_all <- estimated_group_prior_var_all*estimated_group_prior_all*group_size/sample_size #check PVE calculation
#multitissue TWAS analysis with bonferroni adjusted threshold for z scores
load(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag,"_LDR_z_gene.Rd"))
alpha <- 0.05
sig_thresh <- qnorm(1-(alpha/nrow(z_gene)/2), lower=T)
twas_genes <- z_gene$id[abs(z_gene$z)>sig_thresh]
twas_genes_combined <- unique(sapply(twas_genes, function(x){unlist(strsplit(x, "[|]"))[1]}))
n_twas_genes <- length(twas_genes)
n_twas_genes_combined <- length(twas_genes_combined)
n_twas_genes_in_causal <- sum(twas_genes %in% true_genes)
n_twas_genes_in_causal_combined <- sum(twas_genes_combined %in% true_genes_combined)
results_current <- data.frame(simutag=as.character(simutag),
n_causal=as.integer(n_causal),
n_causal_combined=as.integer(n_causal_combined),
n_detected_pip=as.integer(n_ctwas_genes),
n_detected_pip_in_causal=as.integer(n_causal_detected),
n_detected_comb_pip=as.integer(n_ctwas_genes_combined),
n_detected_comb_pip_in_causal=as.integer(n_causal_detected_combined),
pve_snp=as.numeric(rev(estimated_group_pve_all["SNP",])[1]),
pve_weight1=as.numeric(rev(estimated_group_pve_all["Liver",])[1]),
pve_weight2=as.numeric(rev(estimated_group_pve_all["Liver_copy",])[1]),
prior_snp=as.numeric(rev(estimated_group_prior_all["SNP",])[1]),
prior_weight1=as.numeric(rev(estimated_group_prior_all["Liver",])[1]),
prior_weight2=as.numeric(rev(estimated_group_prior_all["Liver_copy",])[1]),
prior_var_snp=as.numeric(rev(estimated_group_prior_var_all["SNP",])[1]),
prior_var_weight1=as.numeric(rev(estimated_group_prior_var_all["Liver",])[1]),
prior_var_weight2=as.numeric(rev(estimated_group_prior_var_all["Liver_copy",])[1]),
n_detected_twas=as.integer(n_twas_genes),
n_detected_twas_in_causal=as.integer(n_twas_genes_in_causal),
n_detected_comb_twas=as.integer(n_twas_genes_combined),
n_detected_comb_twas_in_causal=as.integer(n_twas_genes_in_causal_combined))
results_df <- rbind(results_df, results_current)
}#results using PIP threshold (gene+tissue)
results_df[,c("simutag", "n_causal", "n_detected_pip", "n_detected_pip_in_causal")] simutag n_causal n_detected_pip n_detected_pip_in_causal
1 1-1 118 0 0
2 1-2 95 0 0
3 1-3 116 0 0
4 1-4 117 0 0
5 1-5 113 0 0#mean percent causal using PIP > 0.8
sum(results_df$n_detected_pip_in_causal)/sum(results_df$n_detected_pip)[1] NaN#results using combined PIP threshold
results_df[,c("simutag", "n_causal_combined", "n_detected_comb_pip", "n_detected_comb_pip_in_causal")] simutag n_causal_combined n_detected_comb_pip n_detected_comb_pip_in_causal
1 1-1 118 31 28
2 1-2 95 32 28
3 1-3 116 26 26
4 1-4 117 41 36
5 1-5 113 40 33#mean percent causal using combined PIP > 0.8
sum(results_df$n_detected_comb_pip_in_causal)/sum(results_df$n_detected_comb_pip)[1] 0.8882353#prior inclusion and mean prior inclusion
results_df[,c(which(colnames(results_df)=="simutag"), setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df))))] simutag prior_snp prior_weight1 prior_weight2
1 1-1 0.0002108796 0.005154730 0.005154730
2 1-2 0.0002573964 0.007027990 0.007027990
3 1-3 0.0002761674 0.006451763 0.006451763
4 1-4 0.0002896721 0.012034873 0.012034873
5 1-5 0.0002807151 0.009465010 0.009465010colMeans(results_df[,setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df)))]) prior_snp prior_weight1 prior_weight2
0.0002629661 0.0080268734 0.0080268734 #prior variance and mean prior variance
results_df[,c(which(colnames(results_df)=="simutag"), grep("prior_var", names(results_df)))] simutag prior_var_snp prior_var_weight1 prior_var_weight2
1 1-1 9.443700 32.95046 32.95046
2 1-2 8.605591 22.28596 22.28596
3 1-3 7.982523 26.83210 26.83210
4 1-4 7.486181 14.84881 14.84881
5 1-5 7.210972 21.07834 21.07834colMeans(results_df[,grep("prior_var", names(results_df))]) prior_var_snp prior_var_weight1 prior_var_weight2
8.145793 23.599134 23.599134 #PVE and mean PVE
results_df[,c(which(colnames(results_df)=="simutag"), grep("pve", names(results_df)))] simutag pve_snp pve_weight1 pve_weight2
1 1-1 0.2344277 0.02791214 0.02791214
2 1-2 0.2607447 0.02573879 0.02573879
3 1-3 0.2595045 0.02844846 0.02844846
4 1-4 0.2552697 0.02936695 0.02936695
5 1-5 0.2382823 0.03278560 0.03278560colMeans(results_df[,grep("pve", names(results_df))]) pve_snp pve_weight1 pve_weight2
0.24964577 0.02885039 0.02885039 #TWAS results
results_df[,c(which(colnames(results_df)=="simutag"), grep("twas", names(results_df)))] simutag n_detected_twas n_detected_twas_in_causal n_detected_comb_twas
1 1-1 136 34 68
2 1-2 146 34 73
3 1-3 150 37 75
4 1-4 168 44 84
5 1-5 146 38 73
n_detected_comb_twas_in_causal
1 34
2 34
3 37
4 44
5 38sum(results_df$n_detected_comb_twas_in_causal)/sum(results_df$n_detected_comb_twas)[1] 0.5013405
sessionInfo()R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggpubr_0.6.0 ggplot2_3.4.0 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] tidyselect_1.2.0 xfun_0.35 bslib_0.4.1 purrr_1.0.2
[5] carData_3.0-4 colorspace_2.0-3 vctrs_0.6.3 generics_0.1.3
[9] htmltools_0.5.4 yaml_2.3.6 utf8_1.2.2 rlang_1.1.1
[13] jquerylib_0.1.4 later_1.3.0 pillar_1.8.1 glue_1.6.2
[17] withr_2.5.0 DBI_1.1.3 lifecycle_1.0.3 stringr_1.5.0
[21] munsell_0.5.0 ggsignif_0.6.4 gtable_0.3.1 evaluate_0.19
[25] labeling_0.4.2 knitr_1.41 callr_3.7.3 fastmap_1.1.0
[29] httpuv_1.6.7 ps_1.7.2 fansi_1.0.3 highr_0.9
[33] broom_1.0.2 Rcpp_1.0.9 backports_1.2.1 promises_1.2.0.1
[37] scales_1.2.1 cachem_1.0.6 jsonlite_1.8.4 abind_1.4-5
[41] farver_2.1.0 fs_1.5.2 digest_0.6.31 stringi_1.7.8
[45] rstatix_0.7.2 processx_3.8.0 dplyr_1.0.10 getPass_0.2-2
[49] rprojroot_2.0.3 grid_4.1.0 cli_3.6.1 tools_4.1.0
[53] magrittr_2.0.3 sass_0.4.4 tibble_3.1.8 car_3.1-1
[57] tidyr_1.3.0 whisker_0.4.1 pkgconfig_2.0.3 data.table_1.14.6
[61] assertthat_0.2.1 rmarkdown_2.19 httr_1.4.4 rstudioapi_0.14
[65] R6_2.5.1 git2r_0.30.1 compiler_4.1.0