Enrichment – comparing coloc, twas and ctwas results
XSun
2025-07-11
Last updated: 2025-07-16
Checks: 1 1
Knit directory: multigroup_ctwas_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of
the R Markdown file created these results, you’ll want to first commit
it to the Git repo. If you’re still working on the analysis, you can
ignore this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version ec7934f. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: cv/
Ignored: figures/lz/IBD-ebi-a-GCST004131/single/
Ignored: figures/lz_ld/IBD-ebi-a-GCST004131/single/
Unstaged changes:
Modified: analysis/realdata_final_downstream_enrichment_chatgpt_compare_twasctwascoloc.Rmd
Modified: analysis/realdata_final_downstream_enrichment_compare_twasctwascoloc.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/realdata_final_downstream_enrichment_compare_twasctwascoloc.Rmd)
and HTML
(docs/realdata_final_downstream_enrichment_compare_twasctwascoloc.html)
files. If you’ve configured a remote Git repository (see
?wflow_git_remote), click on the hyperlinks in the table
below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 8d593b2 | XSun | 2025-07-14 | update |
| html | 8d593b2 | XSun | 2025-07-14 | update |
| Rmd | 771aa0d | XSun | 2025-07-11 | update |
| html | 771aa0d | XSun | 2025-07-11 | update |
Introduction
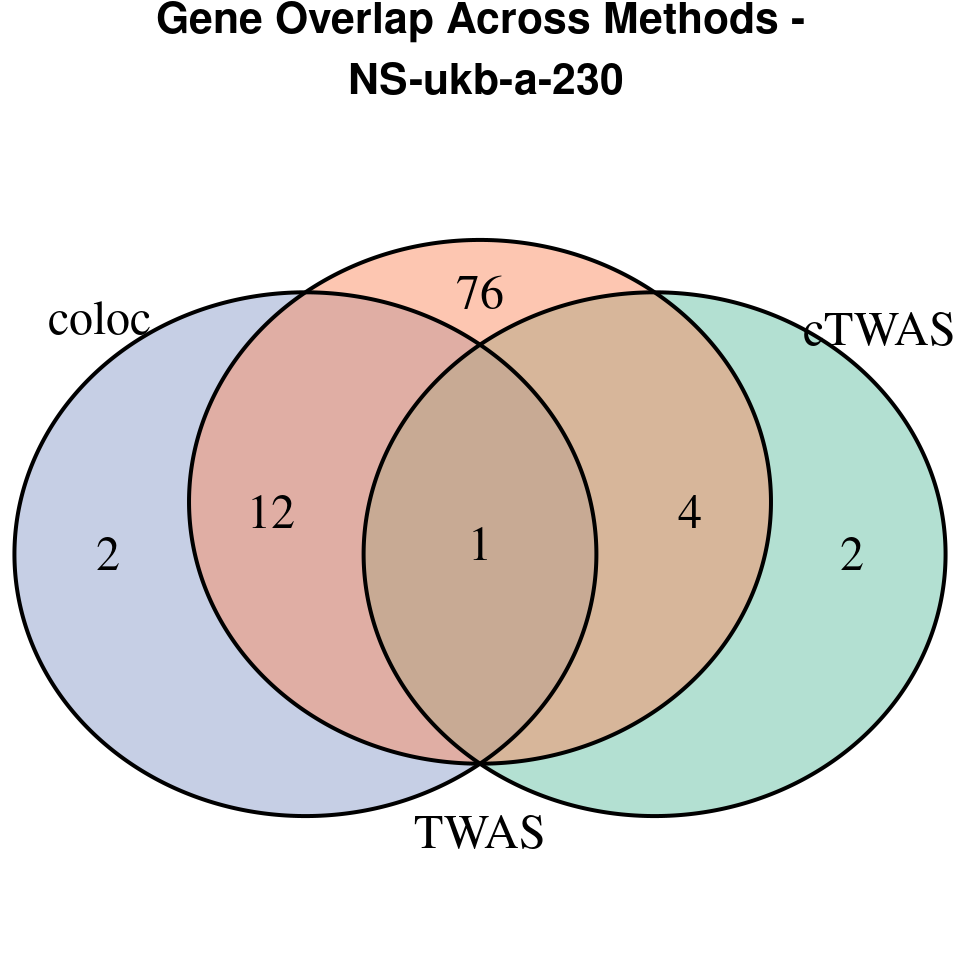
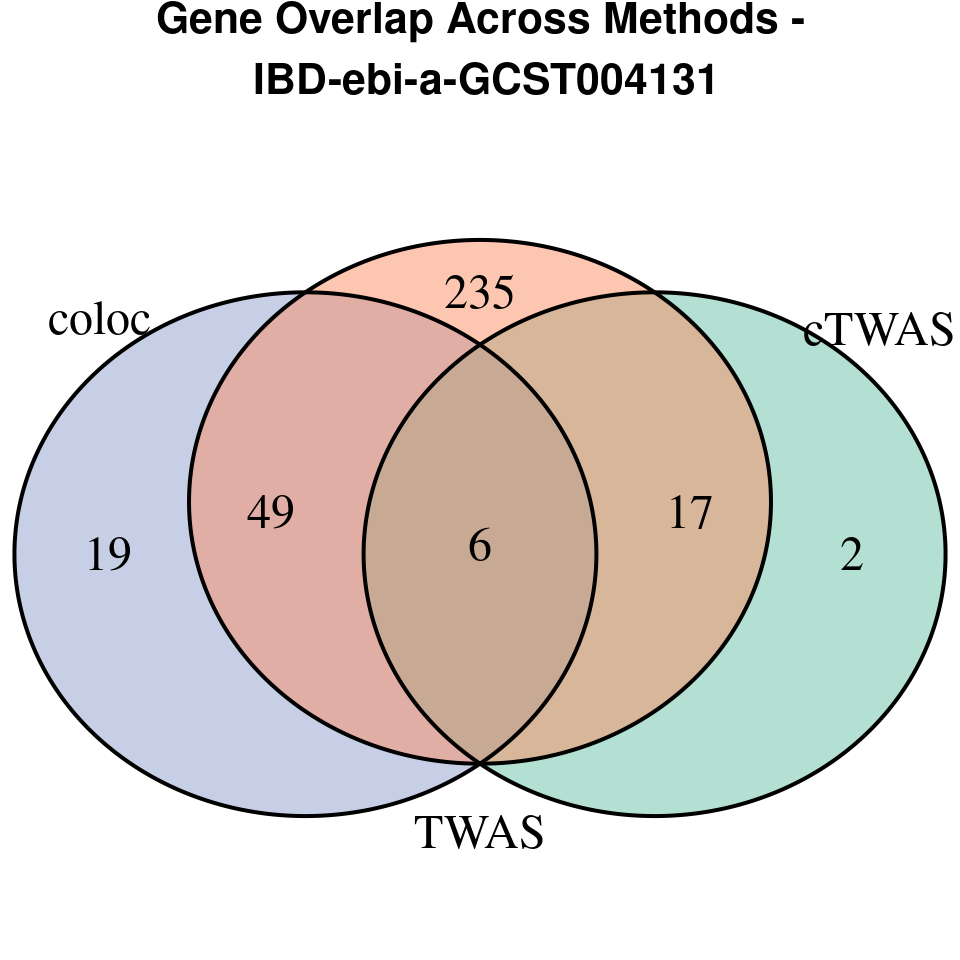
We compared gene set enrichment results (using enrichR) across three analyses: coloc, TWAS, and cTWAS.
- For coloc, we included genes with posterior probability of colocalization (PP4) > 0.8.
- For TWAS, we applied a Bonferroni correction to the TWAS p-values, using a significance threshold calculated as: qnorm(1 - (0.05 / number_of_TWAS_genes / 2), lower.tail = TRUE).
- For cTWAS, we selected genes with a posterior inclusion probability (PIP) > 0.8.
EnrichR only returns the GO terms with
p.adjust < 0.05
Results
DT::datatable(matrix())library(VennDiagram)
trait_nopsy <- c("LDL-ukb-d-30780_irnt","IBD-ebi-a-GCST004131","aFib-ebi-a-GCST006414","SBP-ukb-a-360",
"T1D-GCST90014023","T2D-panukb","BMI-panukb","HB-panukb",
"Height-panukb","HTN-panukb","PLT-panukb","RBC-panukb",
"WBC-ieu-b-30"
)
trait_psy <- c("SCZ-ieu-b-5102","BIP-ieu-b-5110","PD-ieu-b-7","NS-ukb-a-230")
traits <- c(trait_nopsy,trait_psy)
folder_results <- "/project/xinhe/xsun/multi_group_ctwas/23.multi_group_0515/results_downstream/enrichr_compare/"
ctwas_folder_results <- "/project/xinhe/xsun/multi_group_ctwas/23.multi_group_0515/snakemake_outputs/"
for (trait in traits){
print(trait)
ctwas_genes <- readRDS(paste0(ctwas_folder_results,trait,"/",trait,".3qtls.combined_pip_rmmapping_bygroup_final.RDS"))
ctwas_genes <- ctwas_genes$gene_name[ctwas_genes$combined_pip > 0.8]
coloc_genes <- readRDS(paste0(folder_results,trait,".genes.coloc.RDS"))
coloc_genes <- coloc_genes$gene_name[coloc_genes$PP4 > 0.8]
twas_genes <- readRDS(paste0(folder_results,trait,".genes.twas_bonf.RDS"))
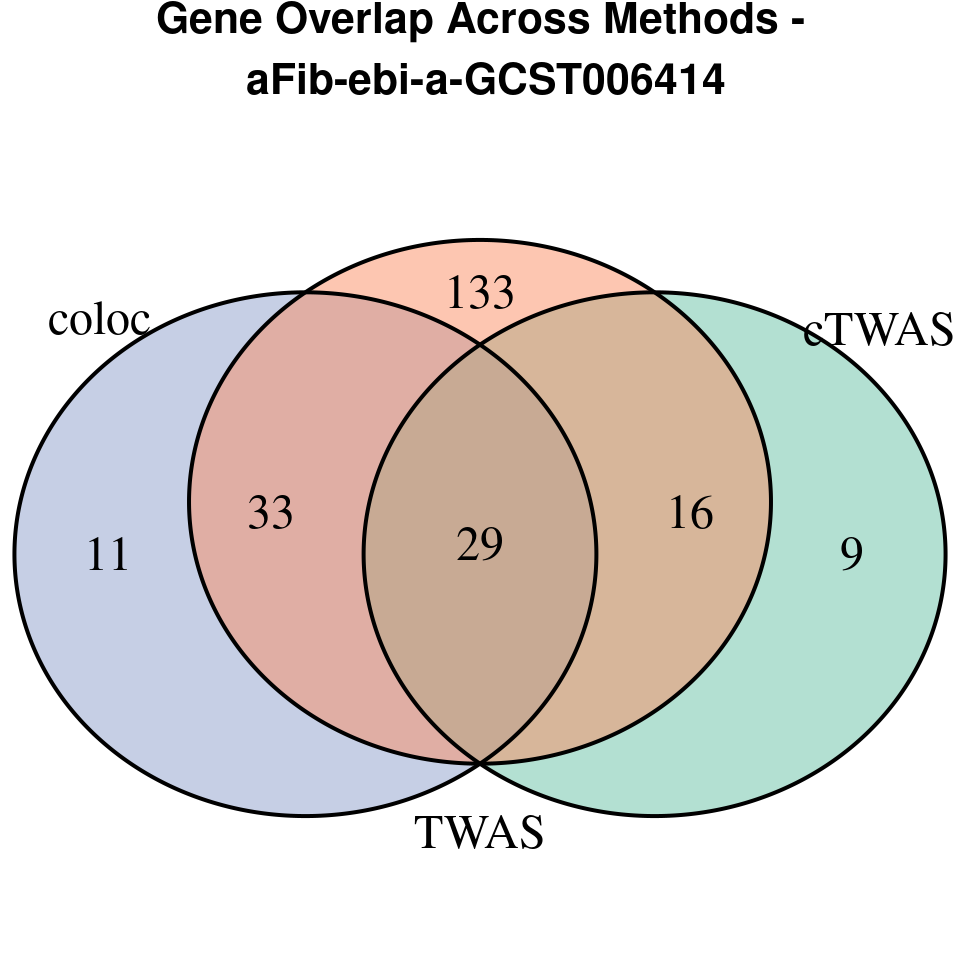
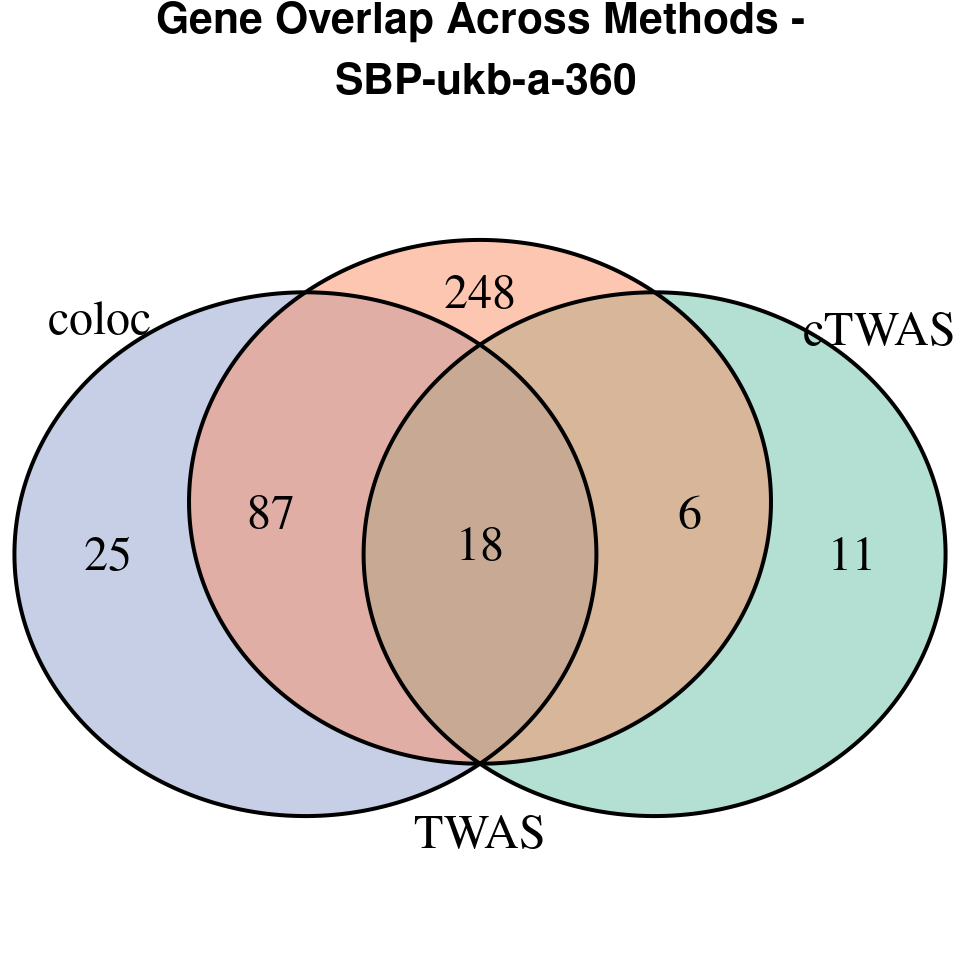
venn.plot <- venn.diagram(
x = list(
cTWAS = ctwas_genes,
TWAS = twas_genes,
coloc = coloc_genes
),
filename = NULL,
output = TRUE,
category.names = c("cTWAS", "TWAS", "coloc"),
cat.cex = 1.5,
cex = 1.5,
print.mode = "raw",
fill = c("#66c2a5", "#fc8d62", "#8da0cb"),
scaled = FALSE
)
grid::grid.newpage()
pushViewport(viewport(layout = grid.layout(10, 1)))
# Add title in row 1
grid.text(paste0("Gene Overlap Across Methods -\n ", trait),
vp = viewport(layout.pos.row = 1),
gp = gpar(fontsize = 16, fontface = "bold"))
# Draw the Venn plot in rows 2–10
pushViewport(viewport(layout.pos.row = 2:10))
grid.draw(venn.plot)
popViewport()
file_ctwas <- paste0(folder_results,trait,".enrichr.ctwas_GO_Biological_Process_2023.RDS")
if(file.exists(file_ctwas)){
df_ctwas <- readRDS(file_ctwas)
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat(knitr::knit_print(DT::datatable(df_ctwas,caption = htmltools::tags$caption( style = 'caption-side: left; text-align: left; color:black; font-size:150% ;',paste0('Enriched GO terms -- ctwas - ',trait)),options = list(pageLength = 2) )))
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
}else{
df_ctwas <- NULL
}
file_twas <- paste0(folder_results,trait,".enrichr.twas_GO_Biological_Process_2023.RDS")
if(file.exists(file_twas)){
df_twas <- readRDS(file_twas)
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat(knitr::knit_print(DT::datatable(df_twas,caption = htmltools::tags$caption( style = 'caption-side: left; text-align: left; color:black; font-size:150% ;',paste0('Enriched GO terms -- twas - ',trait)),options = list(pageLength = 2) )))
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
}else{
df_twas <- NULL
}
file_coloc <- paste0(folder_results,trait,".enrichr.coloc_GO_Biological_Process_2023.RDS")
if(file.exists(file_coloc)){
df_coloc <- readRDS(file_coloc)
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat(knitr::knit_print(DT::datatable(df_coloc,caption = htmltools::tags$caption( style = 'caption-side: left; text-align: left; color:black; font-size:150% ;',paste0('Enriched GO terms -- coloc - ',trait)),options = list(pageLength = 2) )))
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
cat("<br>")
}else{
df_coloc <- NULL
}
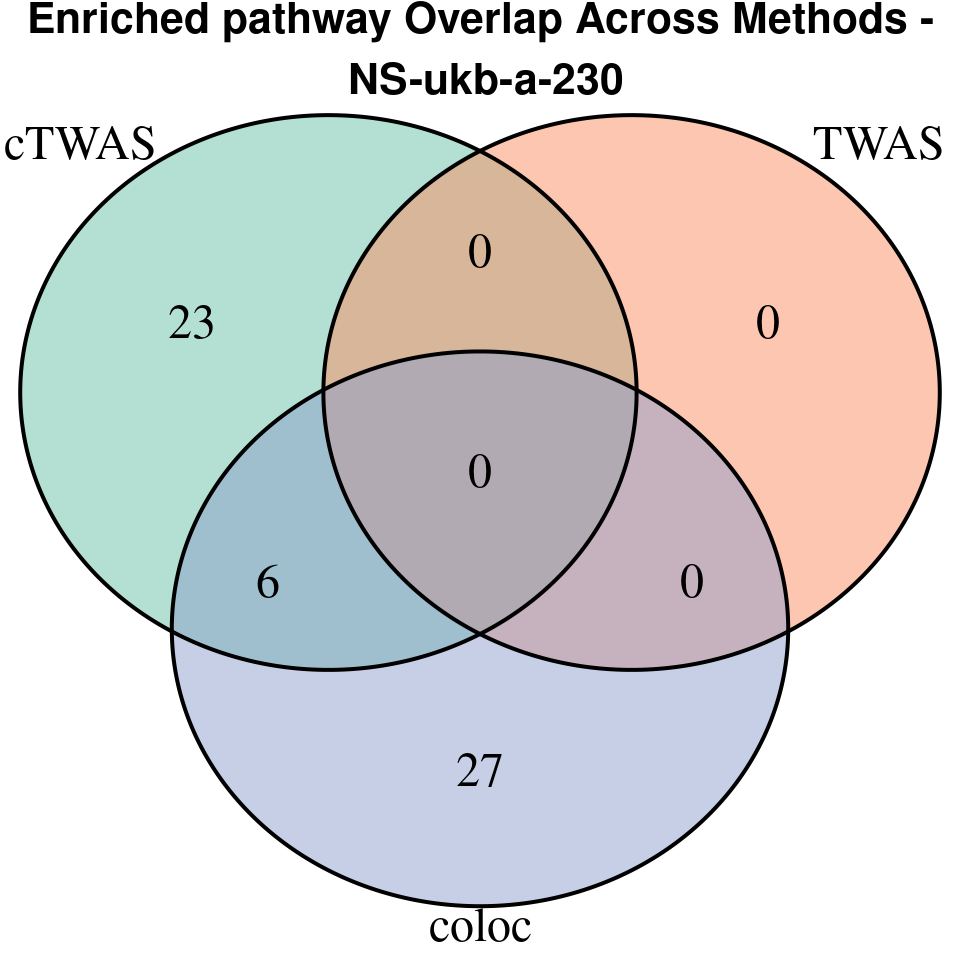
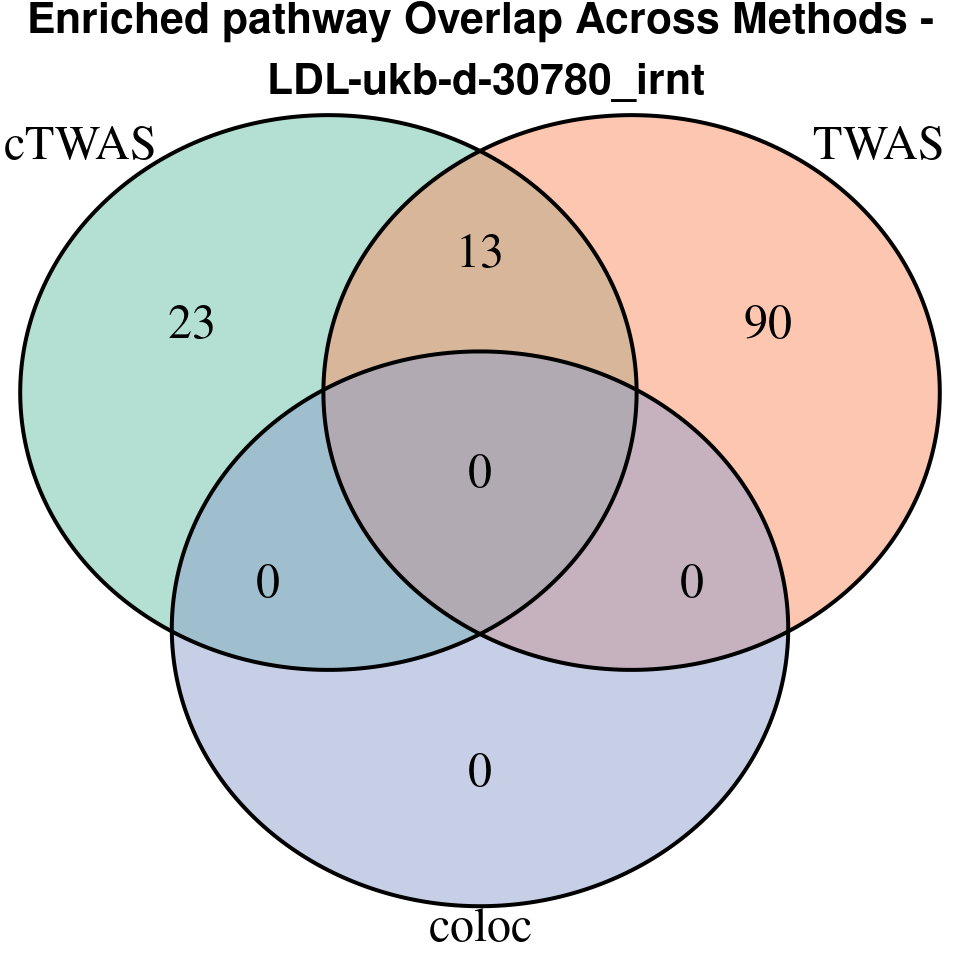
terms_ctwas <- if (!is.null(df_ctwas)) df_ctwas$Term else character(0)
terms_twas <- if (!is.null(df_twas)) df_twas$Term else character(0)
terms_coloc <- if (!is.null(df_coloc)) df_coloc$Term else character(0)
# Create named list
term_lists <- list(
cTWAS = terms_ctwas,
TWAS = terms_twas,
coloc = terms_coloc
)
# Create Venn diagram
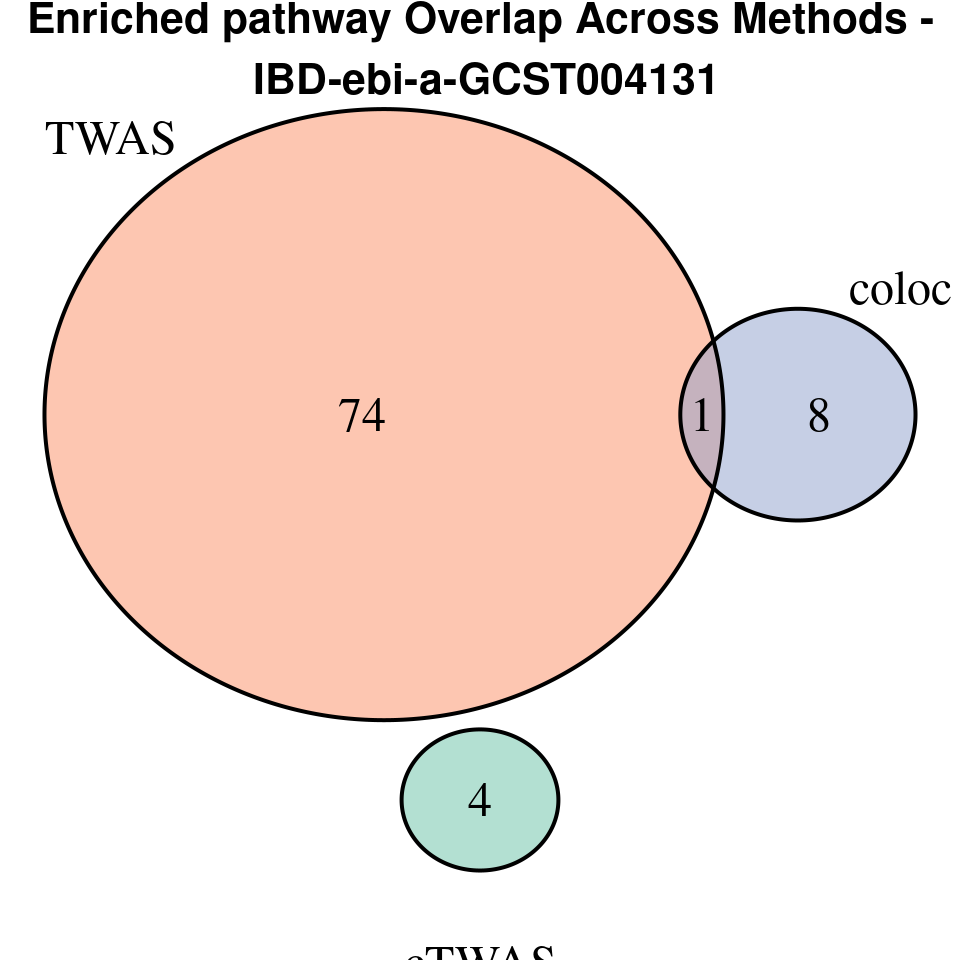
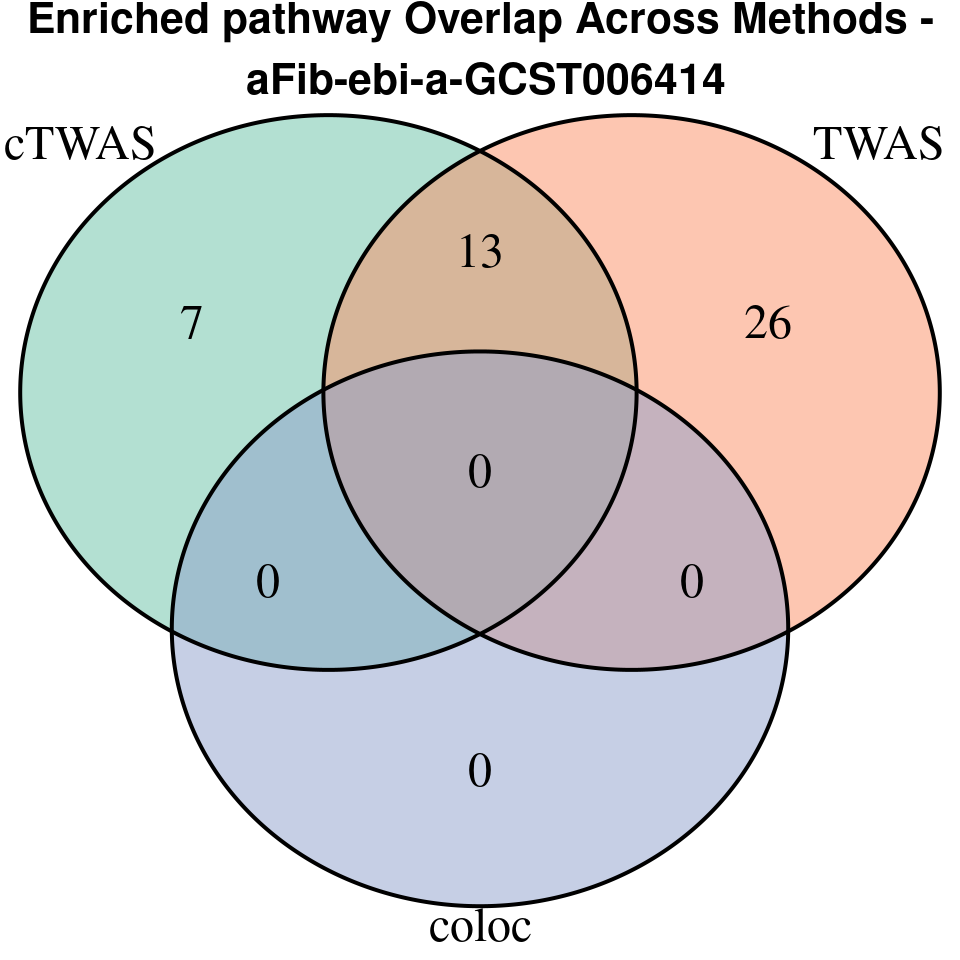
venn.plot <- venn.diagram(
x = term_lists,
filename = NULL,
output = TRUE,
category.names = c("cTWAS", "TWAS", "coloc"),
cat.cex = 1.5,
cex = 1.5,
print.mode = "raw",
fill = c("#66c2a5", "#fc8d62", "#8da0cb"),
scaled = FALSE
)
# Plot with title
grid.newpage()
pushViewport(viewport(layout = grid.layout(10, 1)))
grid.text(paste0("Enriched pathway Overlap Across Methods -\n ", trait),
vp = viewport(layout.pos.row = 1),
gp = gpar(fontsize = 16, fontface = "bold"))
pushViewport(viewport(layout.pos.row = 2:10))
grid.draw(venn.plot)
popViewport()
}[1] “LDL-ukb-d-30780_irnt”

| Version | Author | Date |
|---|---|---|
| 771aa0d | XSun | 2025-07-11 |
| Version | Author | Date |
|---|---|---|
| 771aa0d | XSun | 2025-07-11 |
[1] “IBD-ebi-a-GCST004131”
| Version | Author | Date |
|---|---|---|
| 771aa0d | XSun | 2025-07-11 |
| Version | Author | Date |
|---|---|---|
| 771aa0d | XSun | 2025-07-11 |
[1] “aFib-ebi-a-GCST006414”
| Version | Author | Date |
|---|---|---|
| 771aa0d | XSun | 2025-07-11 |
| Version | Author | Date |
|---|---|---|
| 771aa0d | XSun | 2025-07-11 |
[1] “SBP-ukb-a-360”
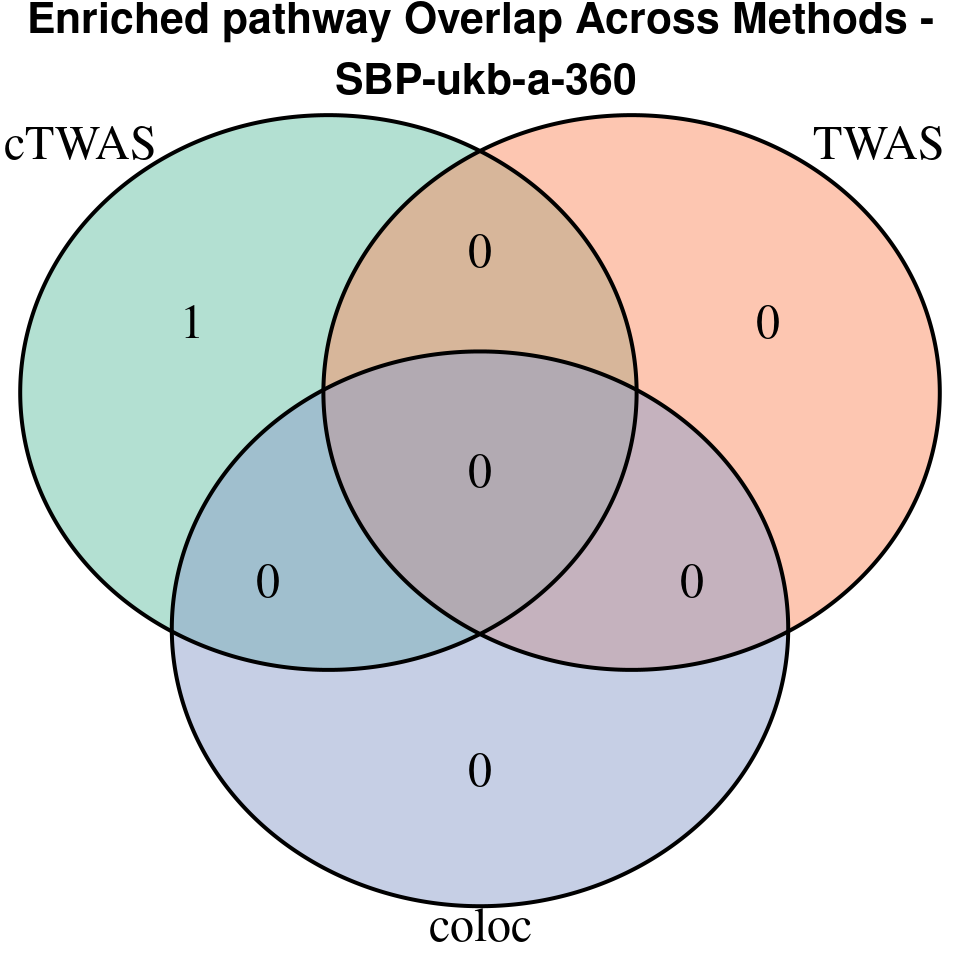
[1] “T1D-GCST90014023”
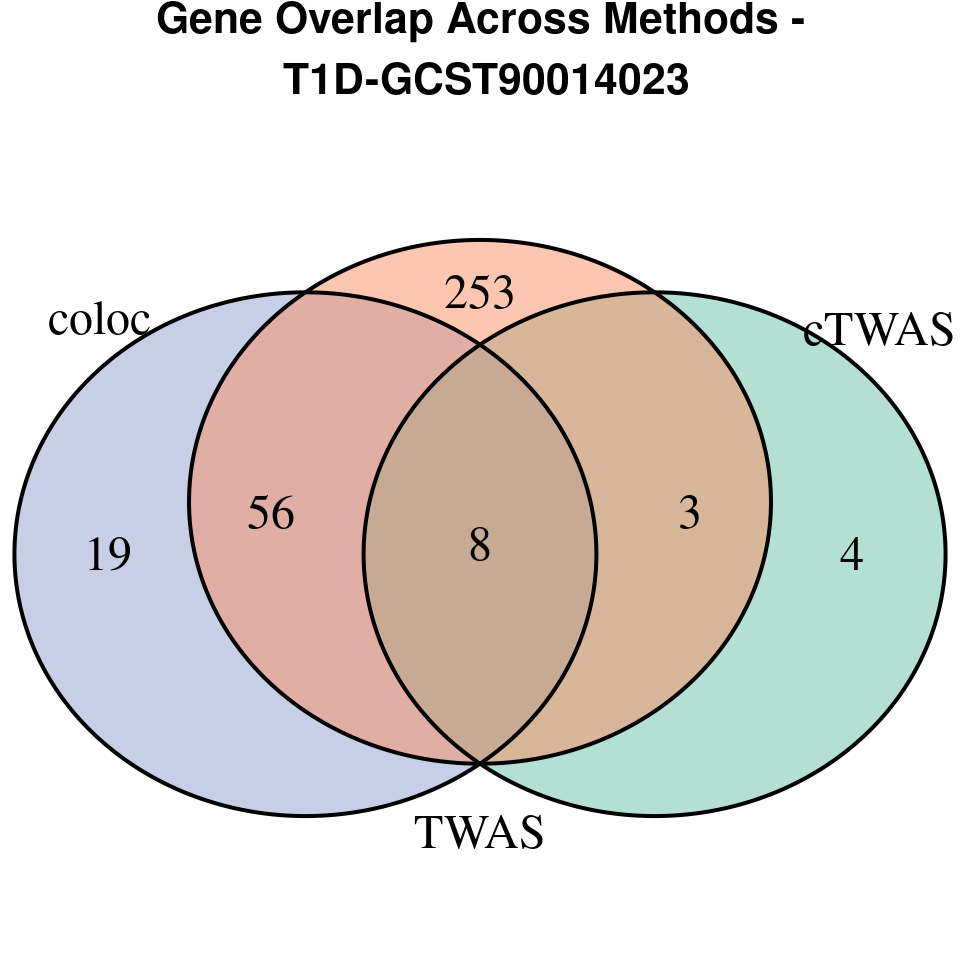
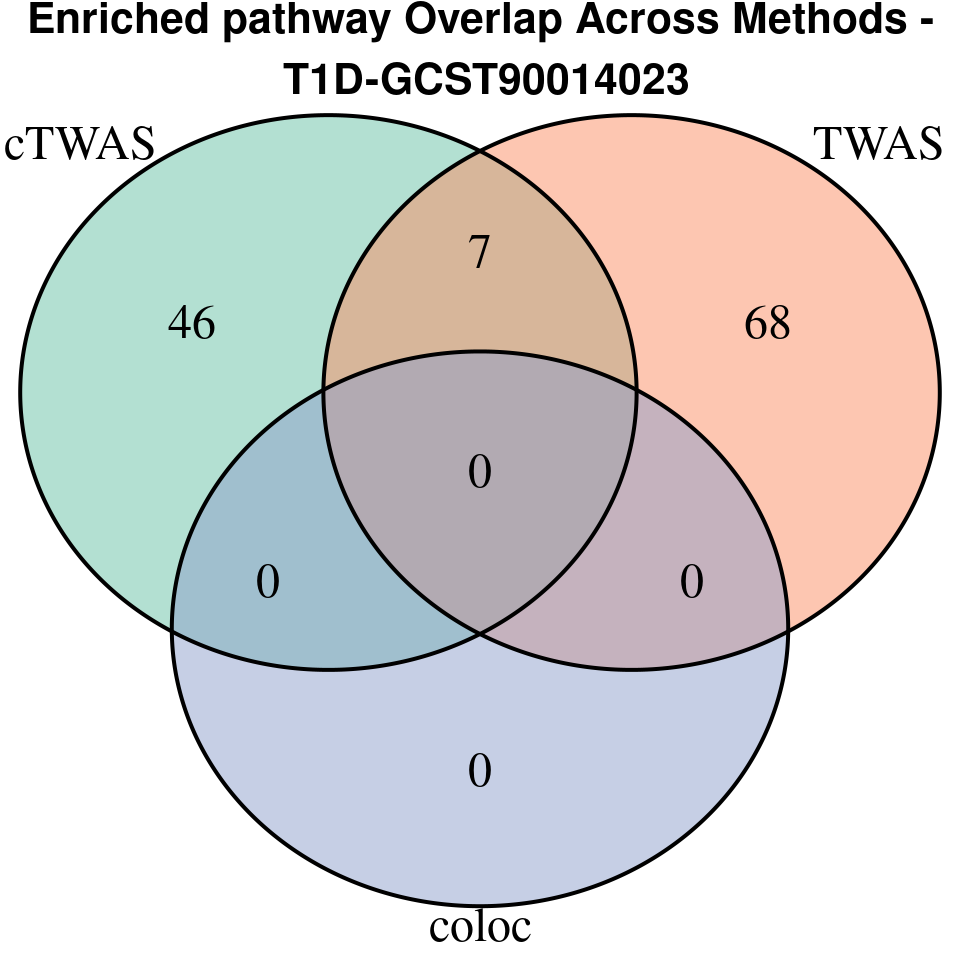
[1] “T2D-panukb”
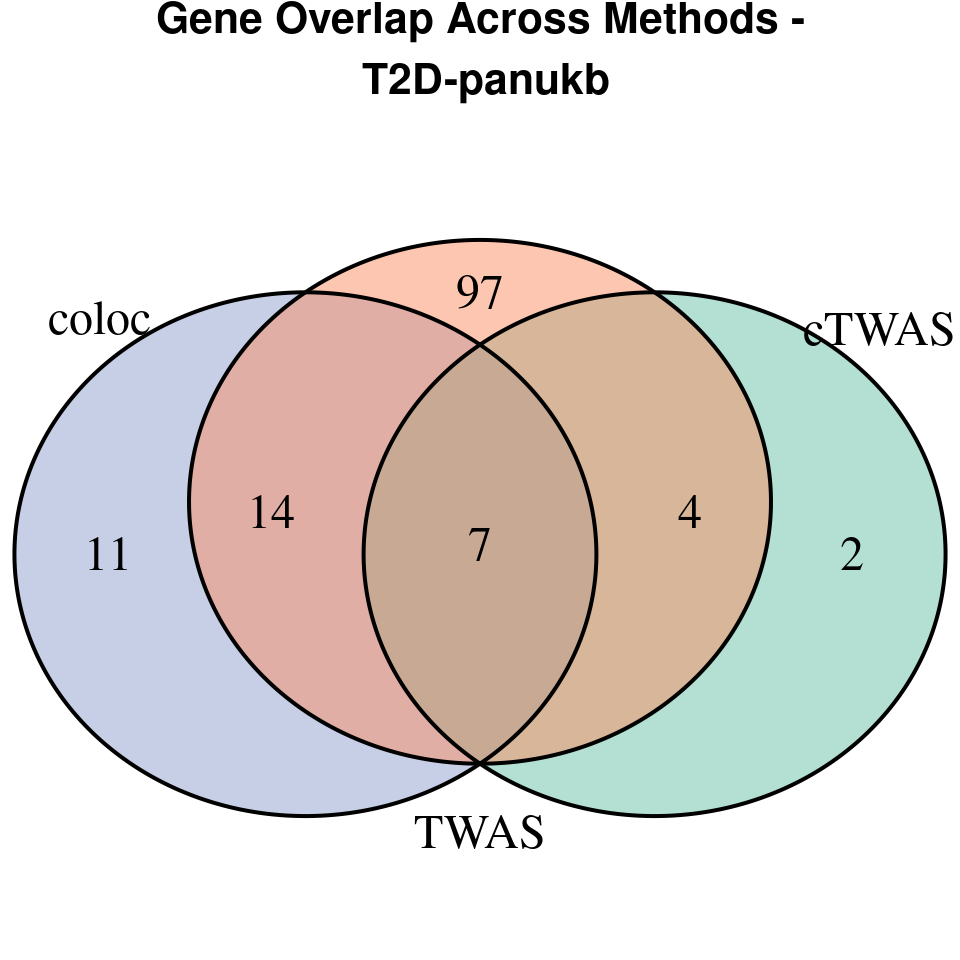
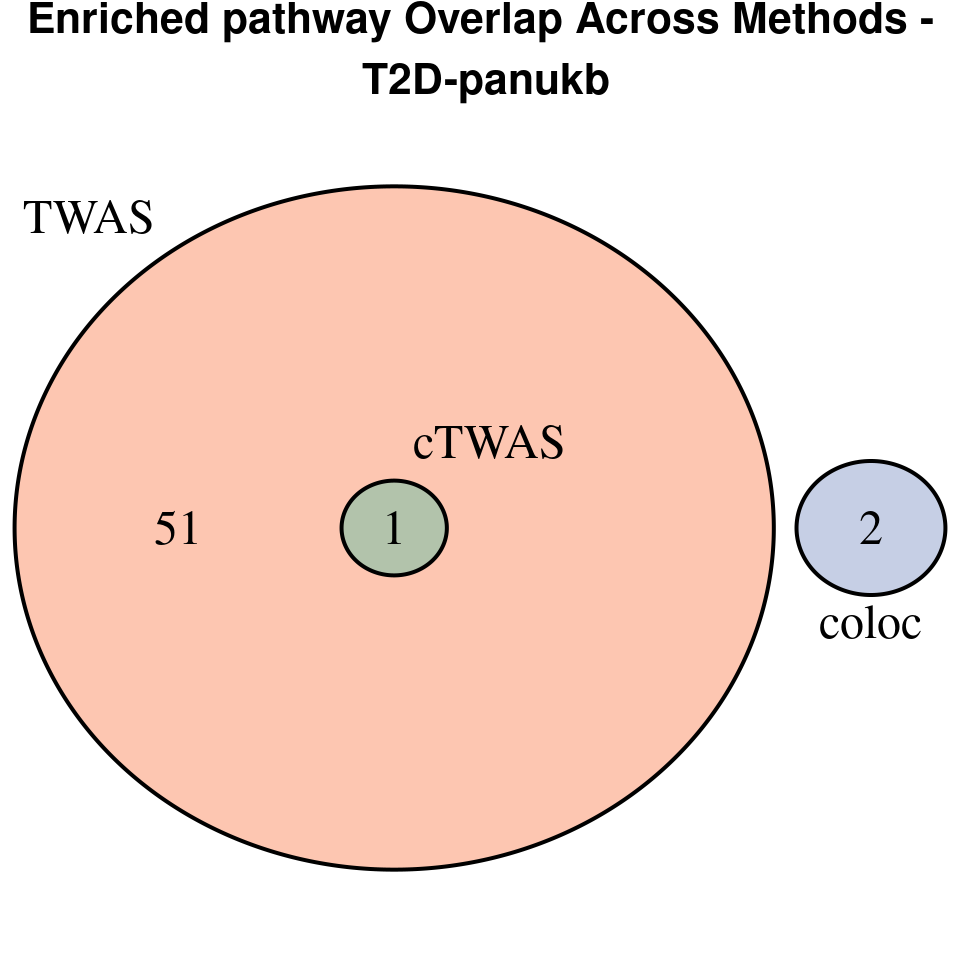
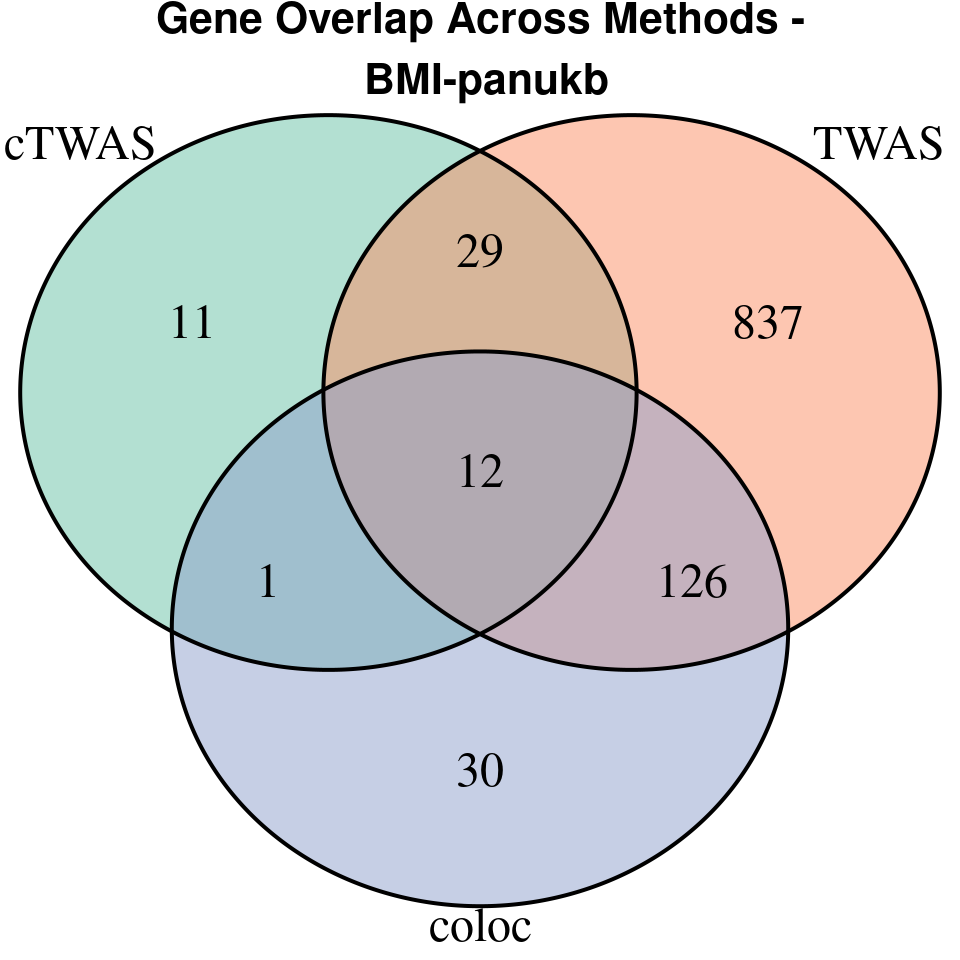
[1] “BMI-panukb”
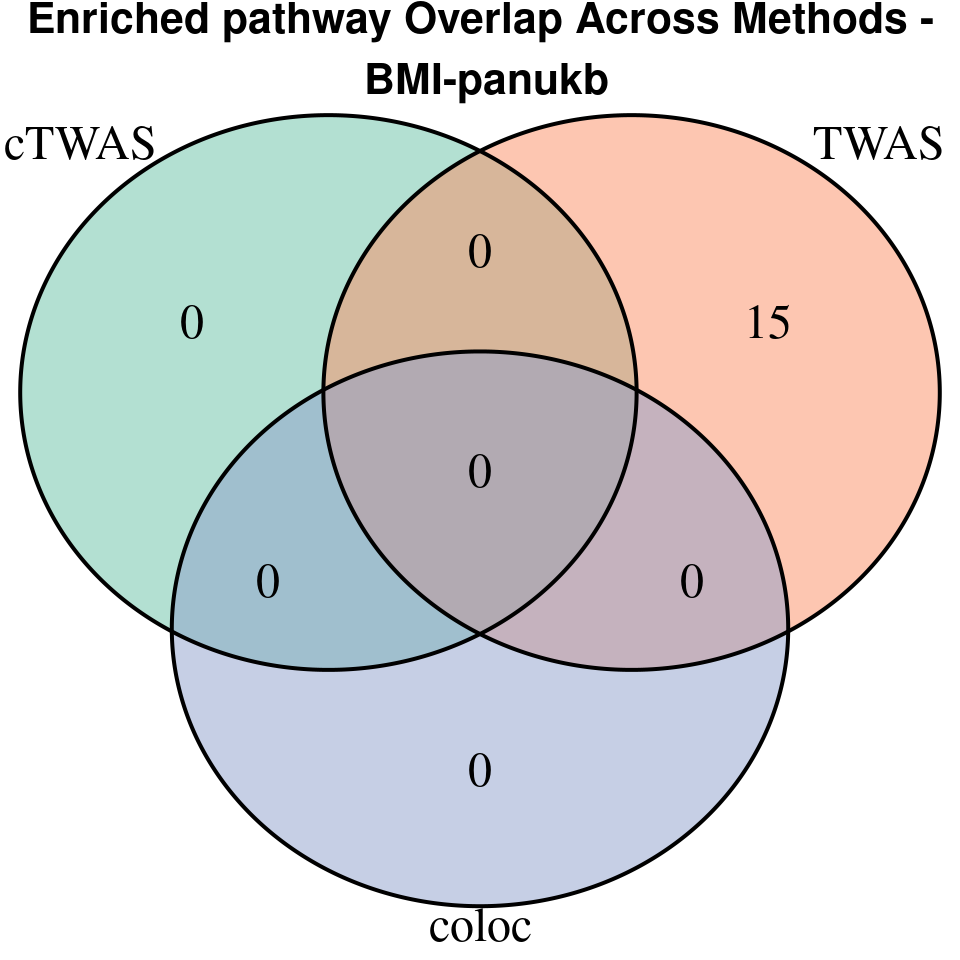
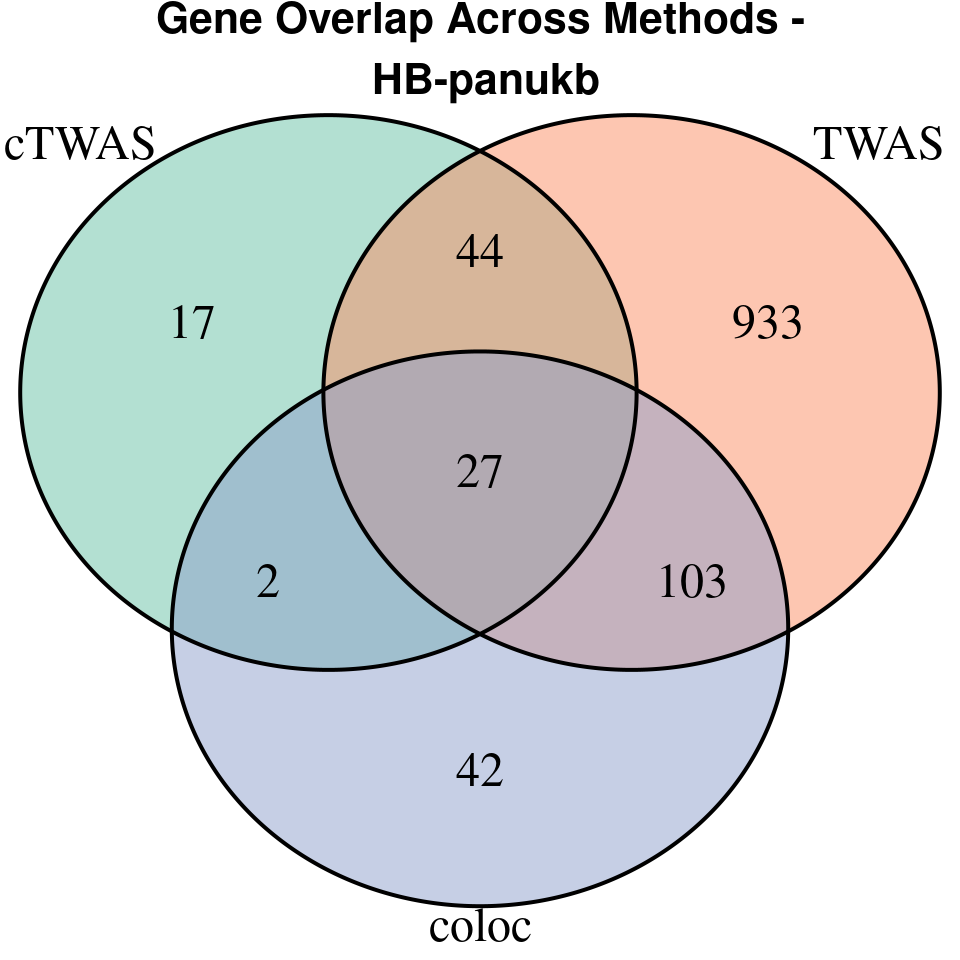
[1] “HB-panukb”
| Version | Author | Date |
|---|---|---|
| 8d593b2 | XSun | 2025-07-14 |
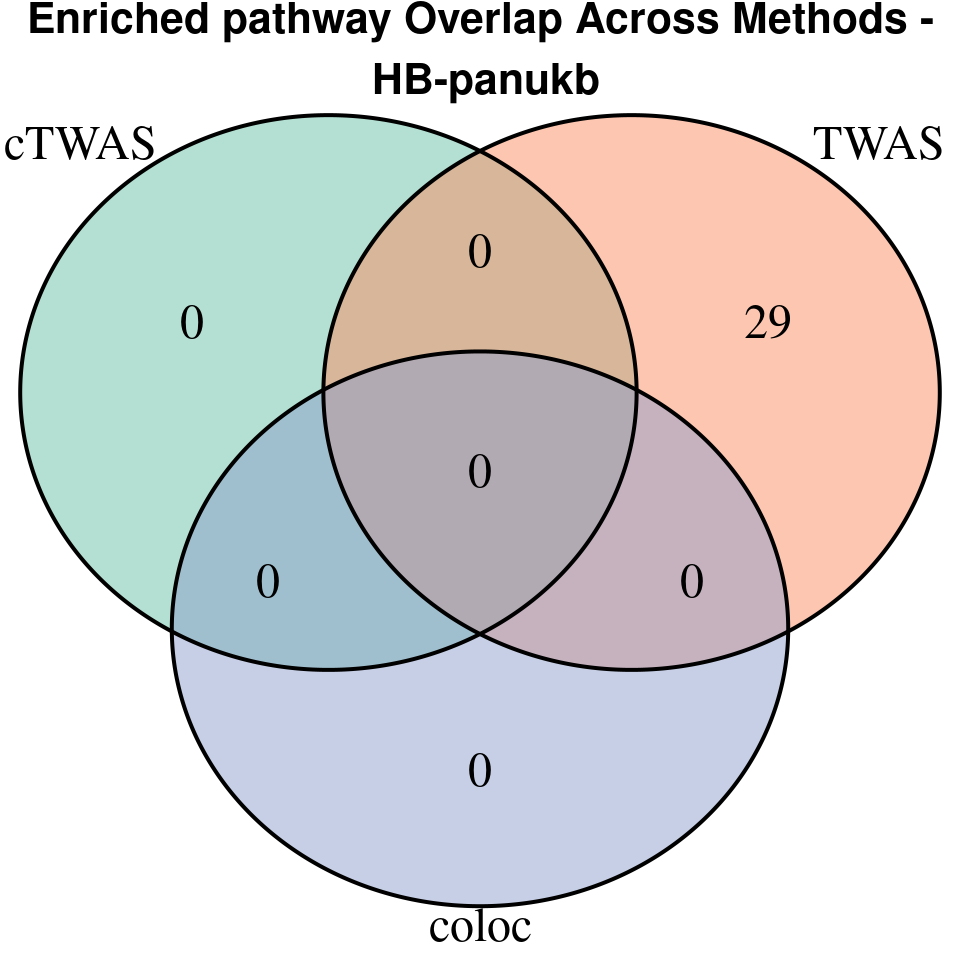
| Version | Author | Date |
|---|---|---|
| 8d593b2 | XSun | 2025-07-14 |
[1] “Height-panukb”

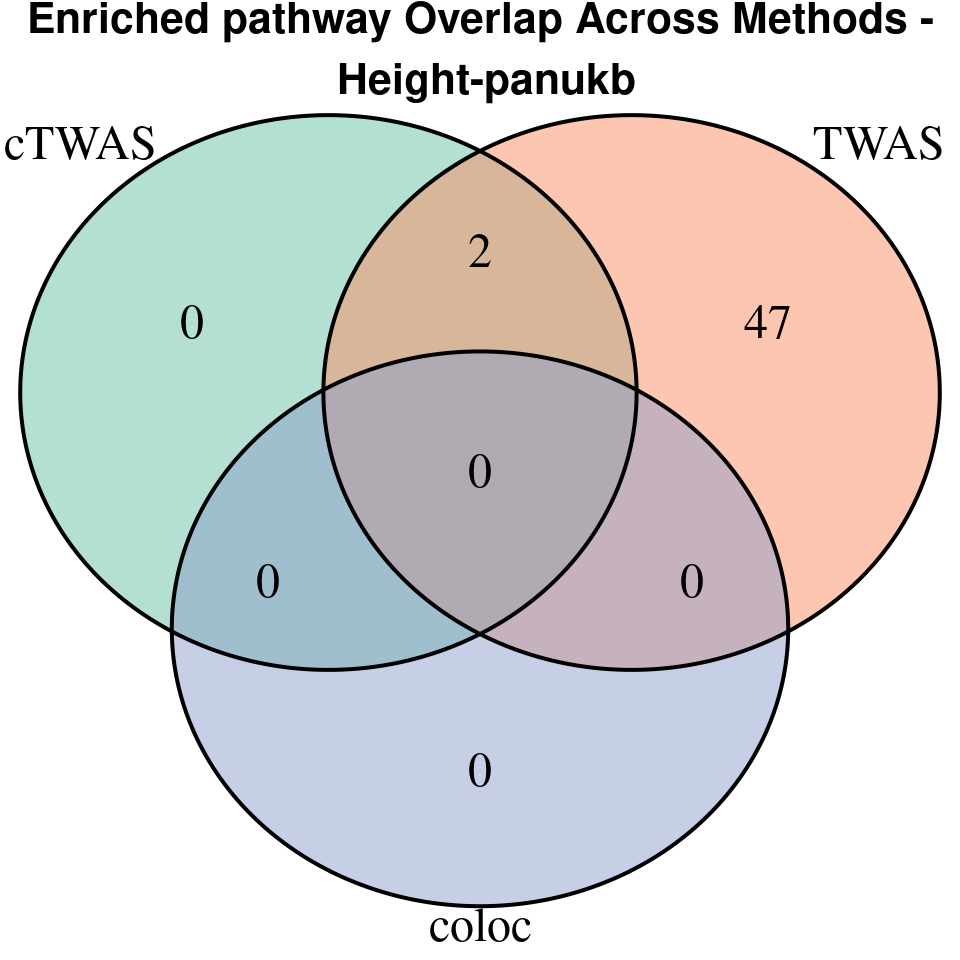 [1]
“HTN-panukb”
[1]
“HTN-panukb”
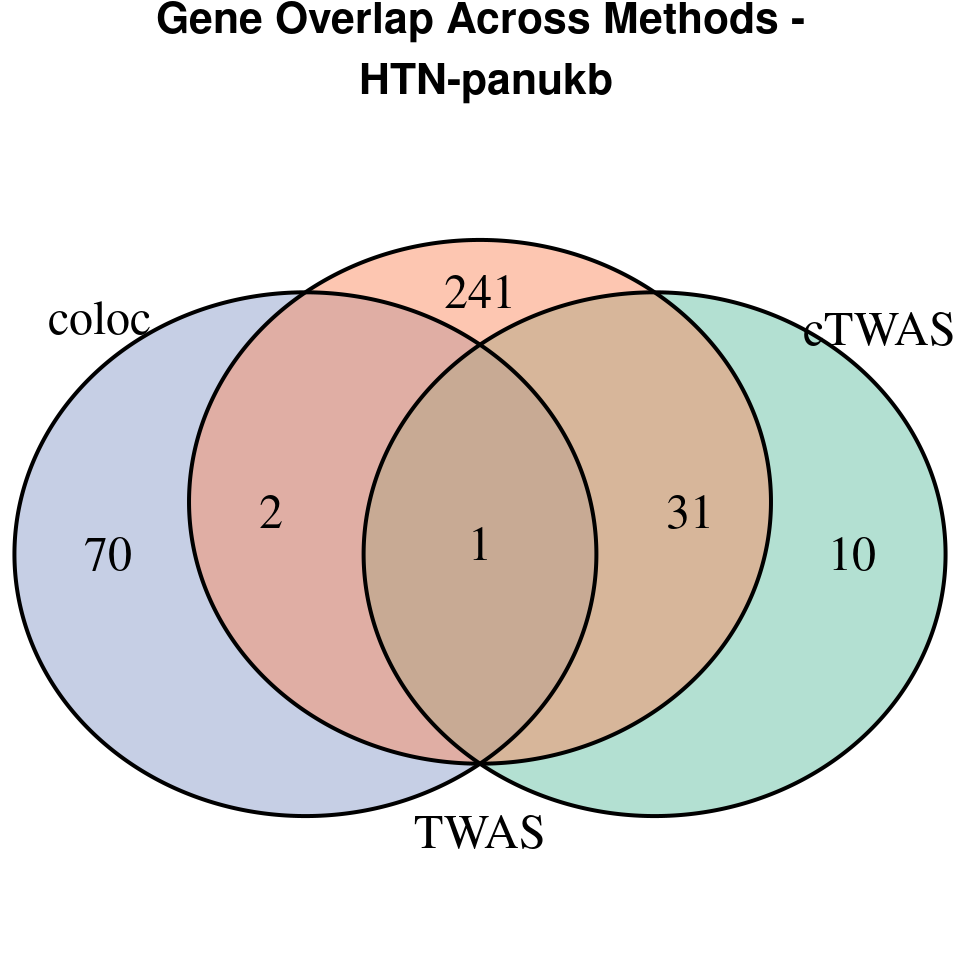
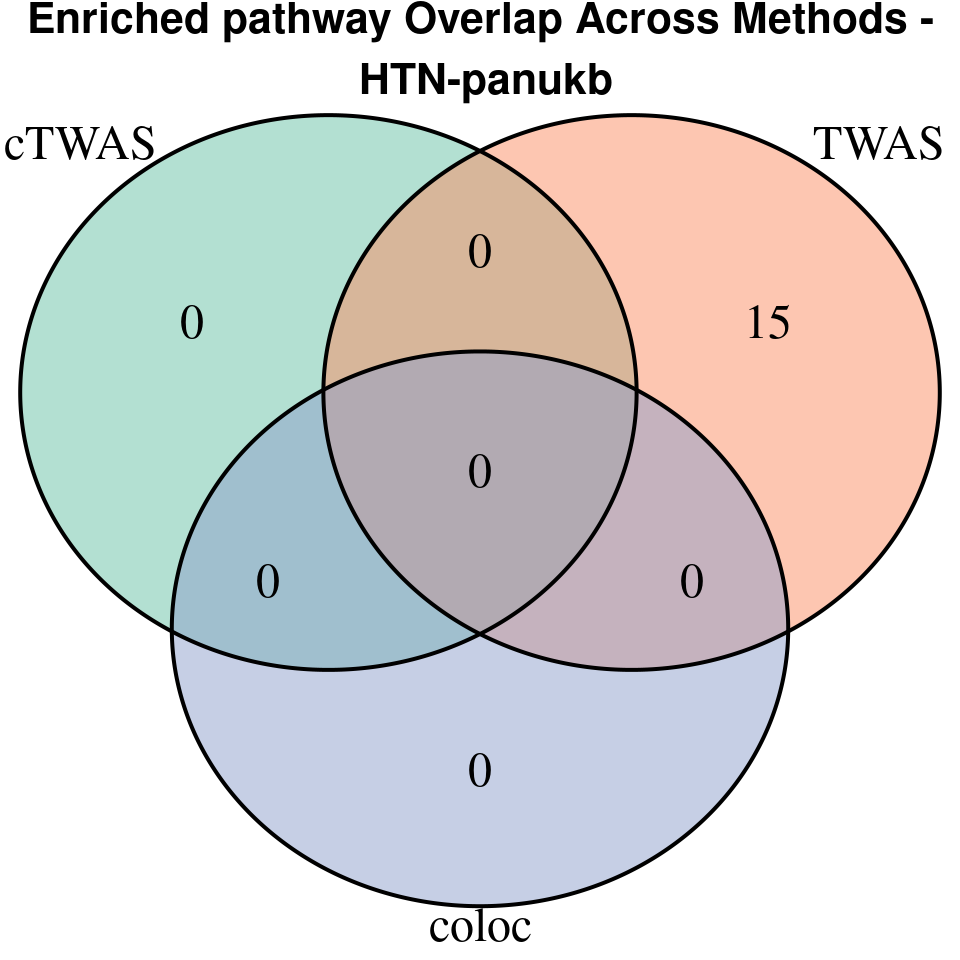 [1]
“PLT-panukb”
[1]
“PLT-panukb”
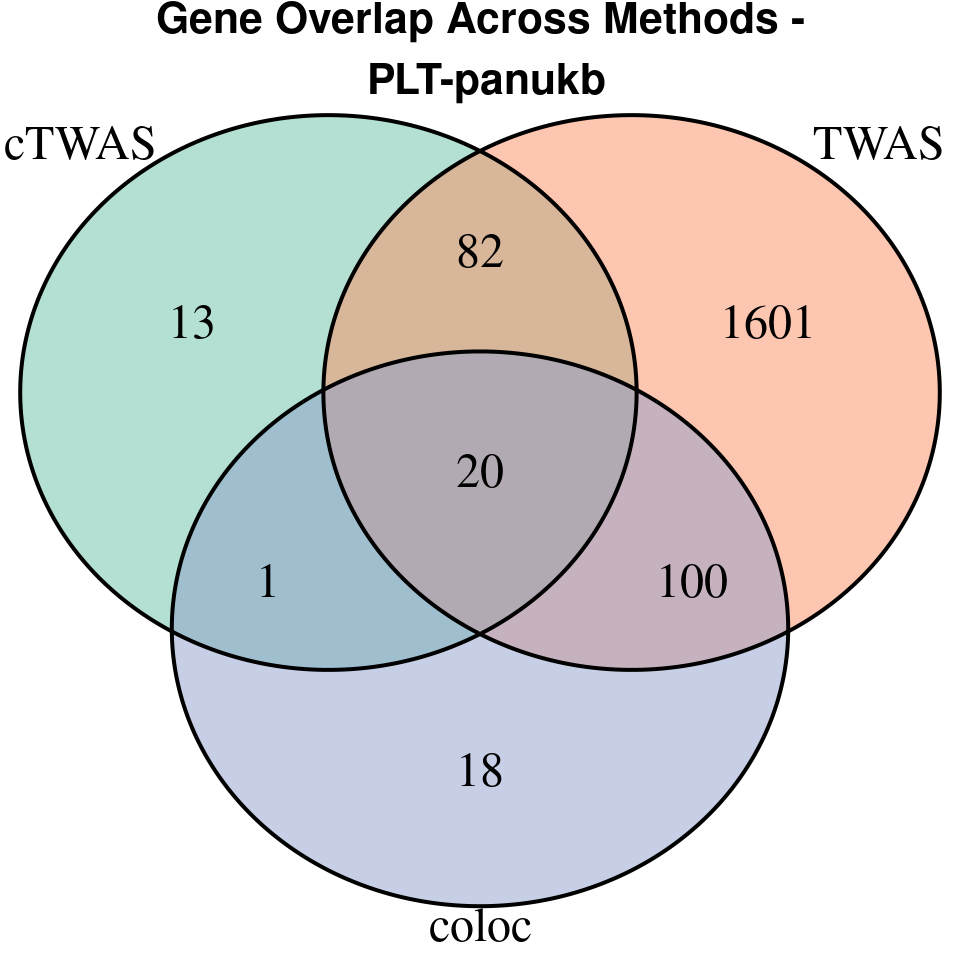
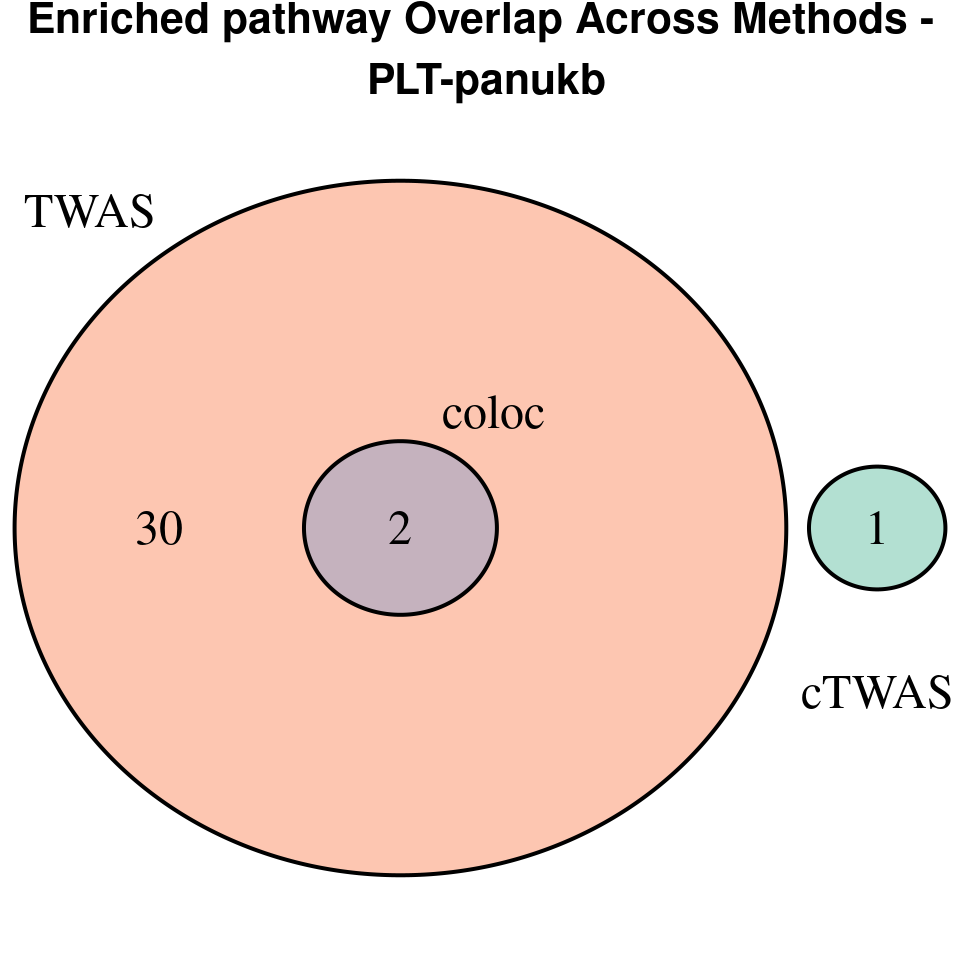 [1]
“RBC-panukb”
[1]
“RBC-panukb”
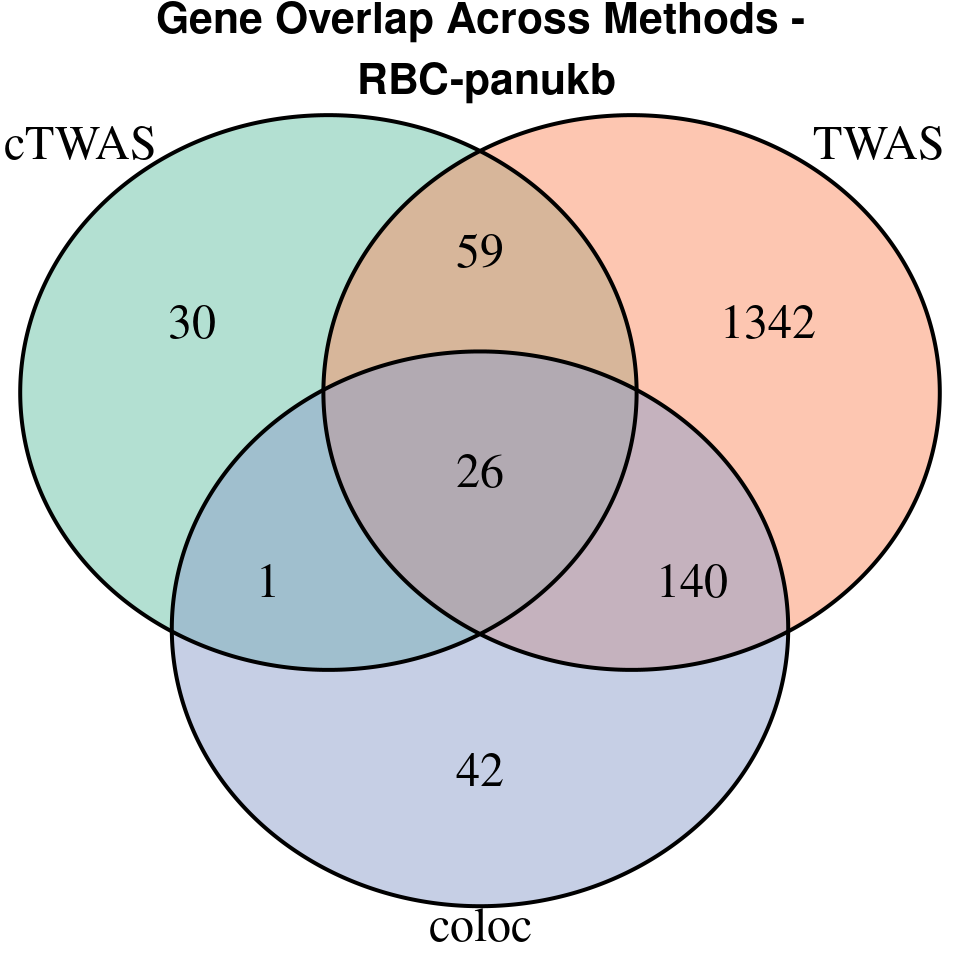
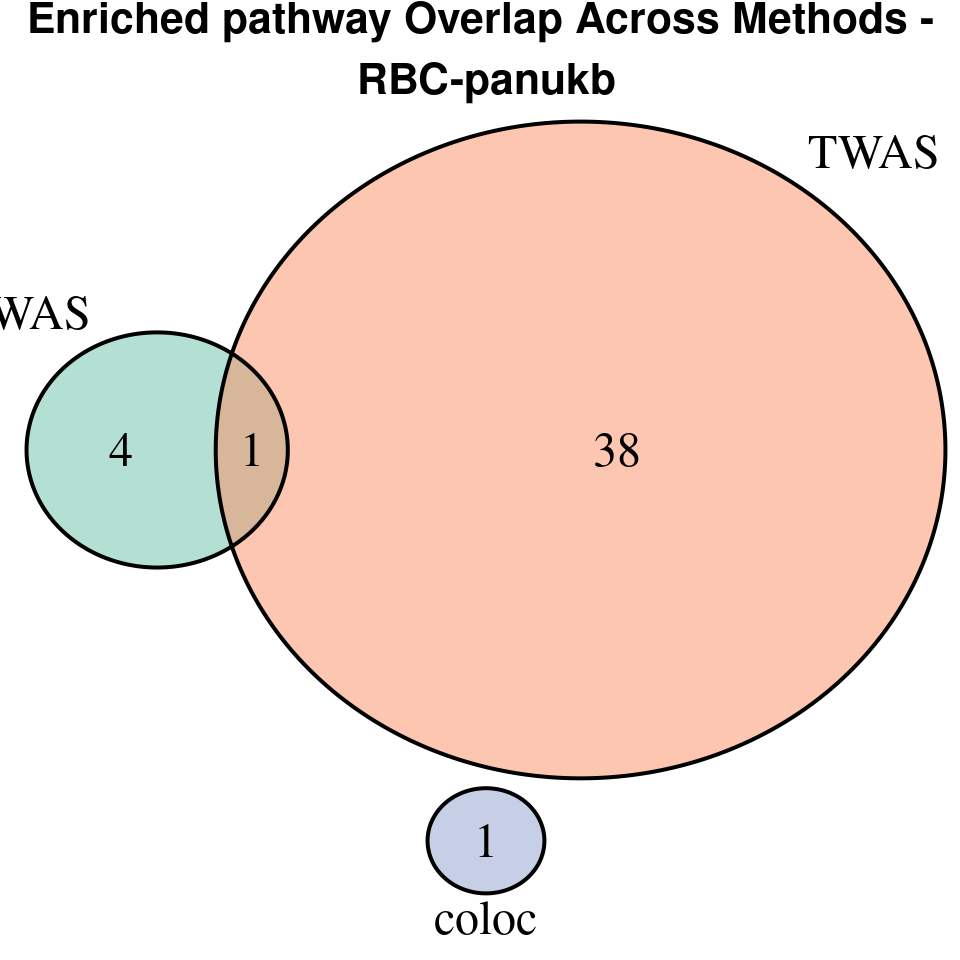 [1]
“WBC-ieu-b-30”
[1]
“WBC-ieu-b-30”
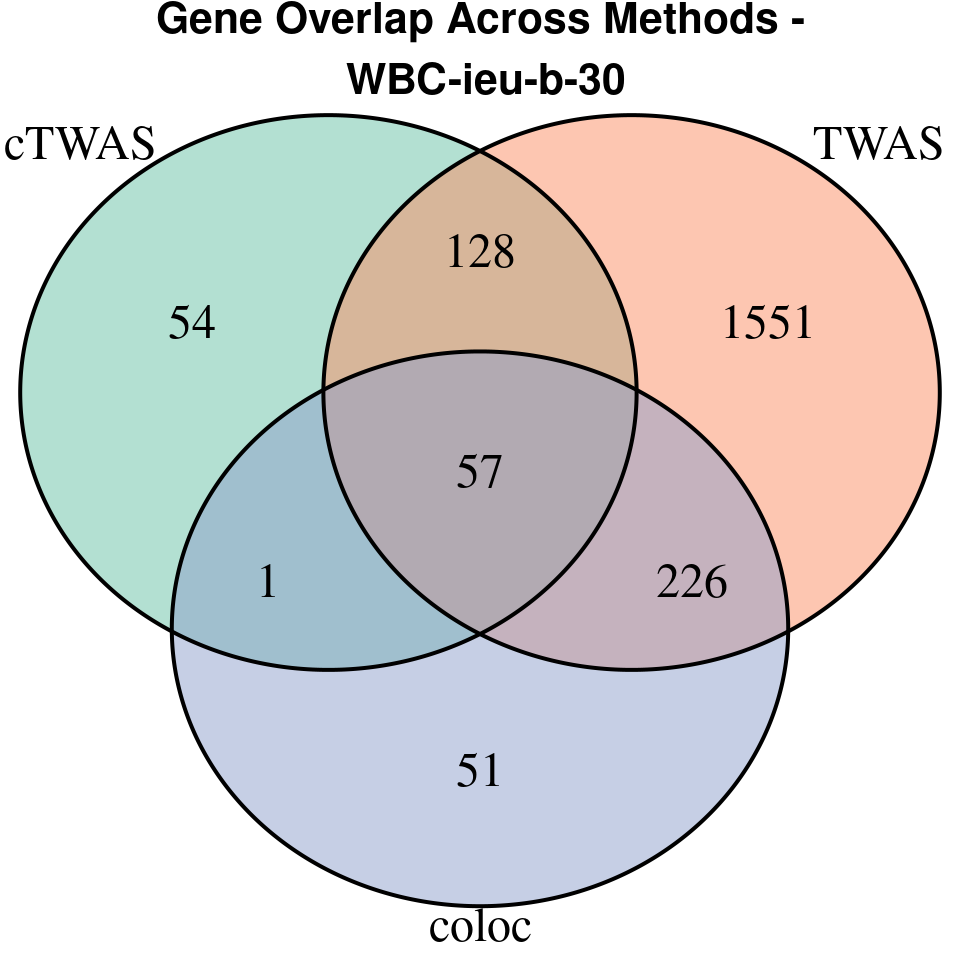
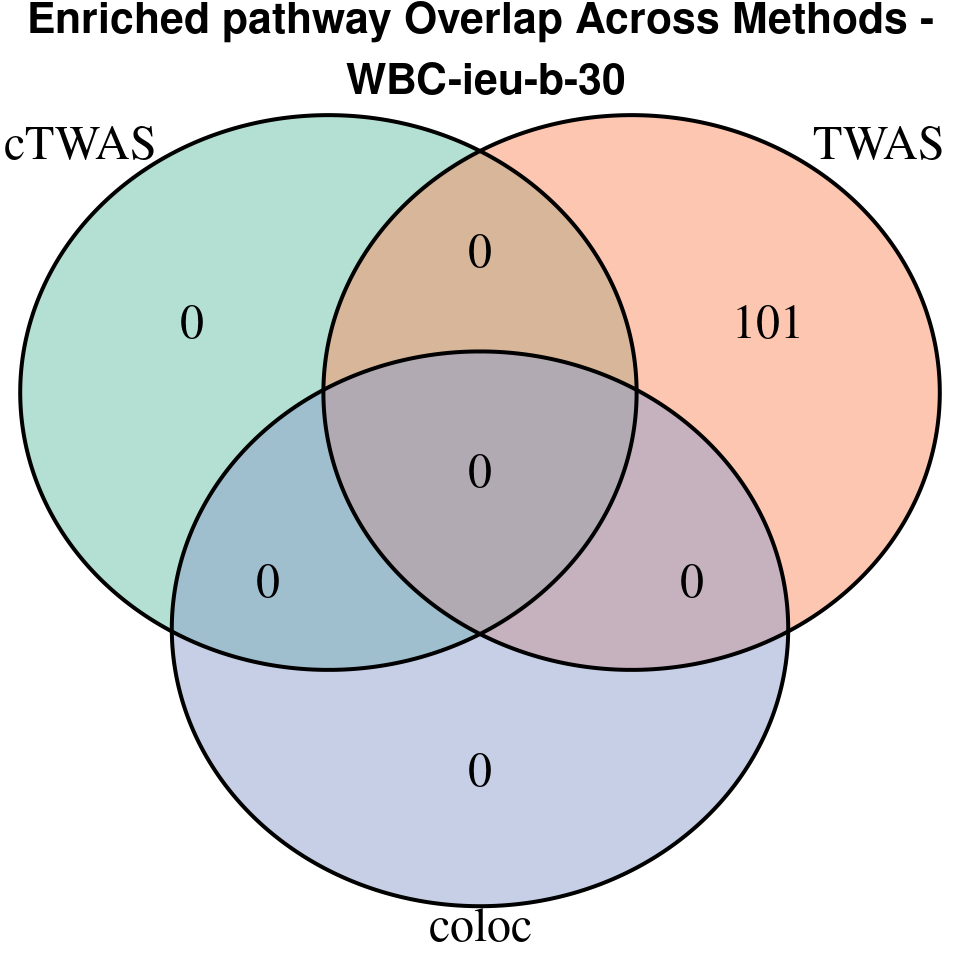 [1]
“SCZ-ieu-b-5102”
[1]
“SCZ-ieu-b-5102”
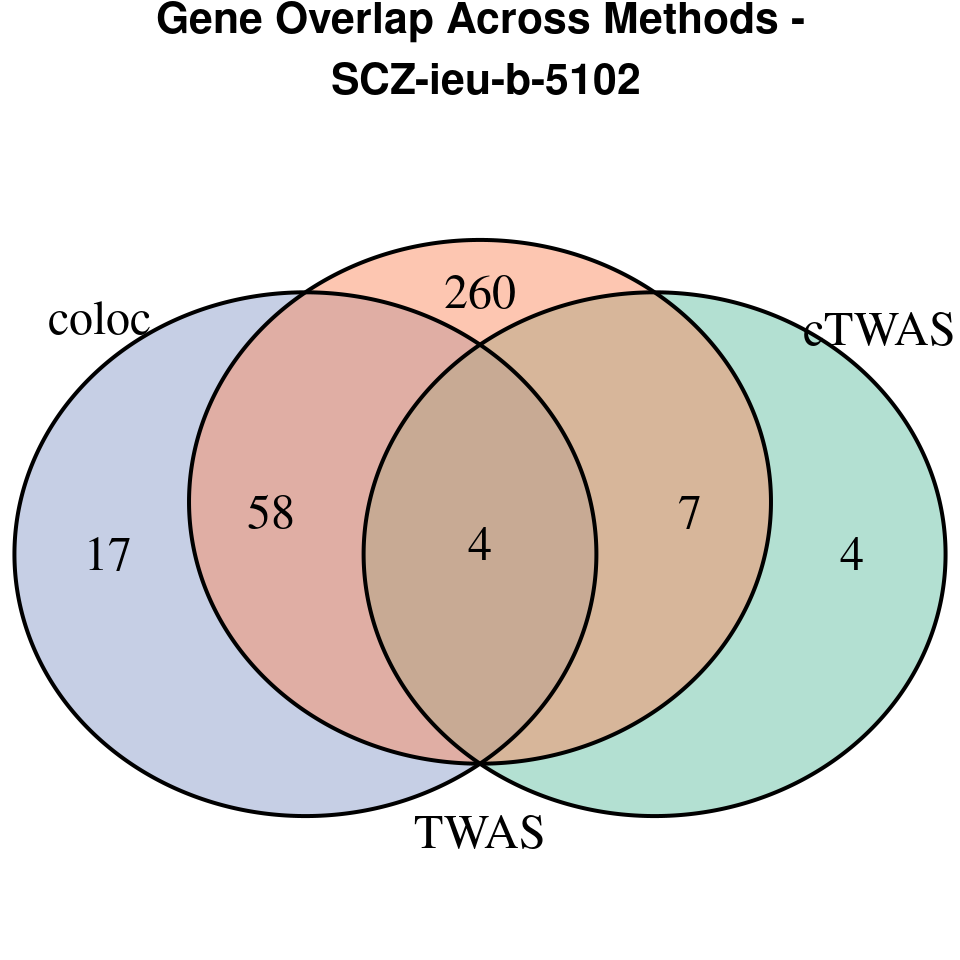
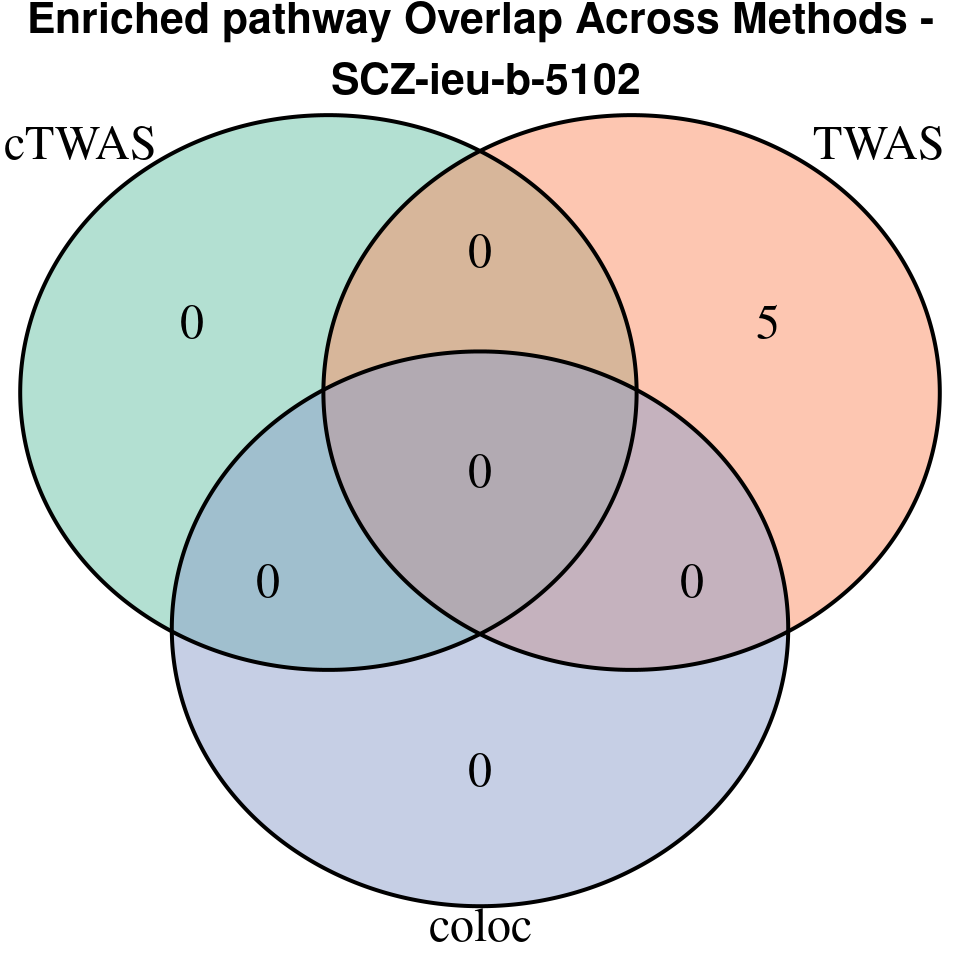 [1]
“BIP-ieu-b-5110”
[1]
“BIP-ieu-b-5110”
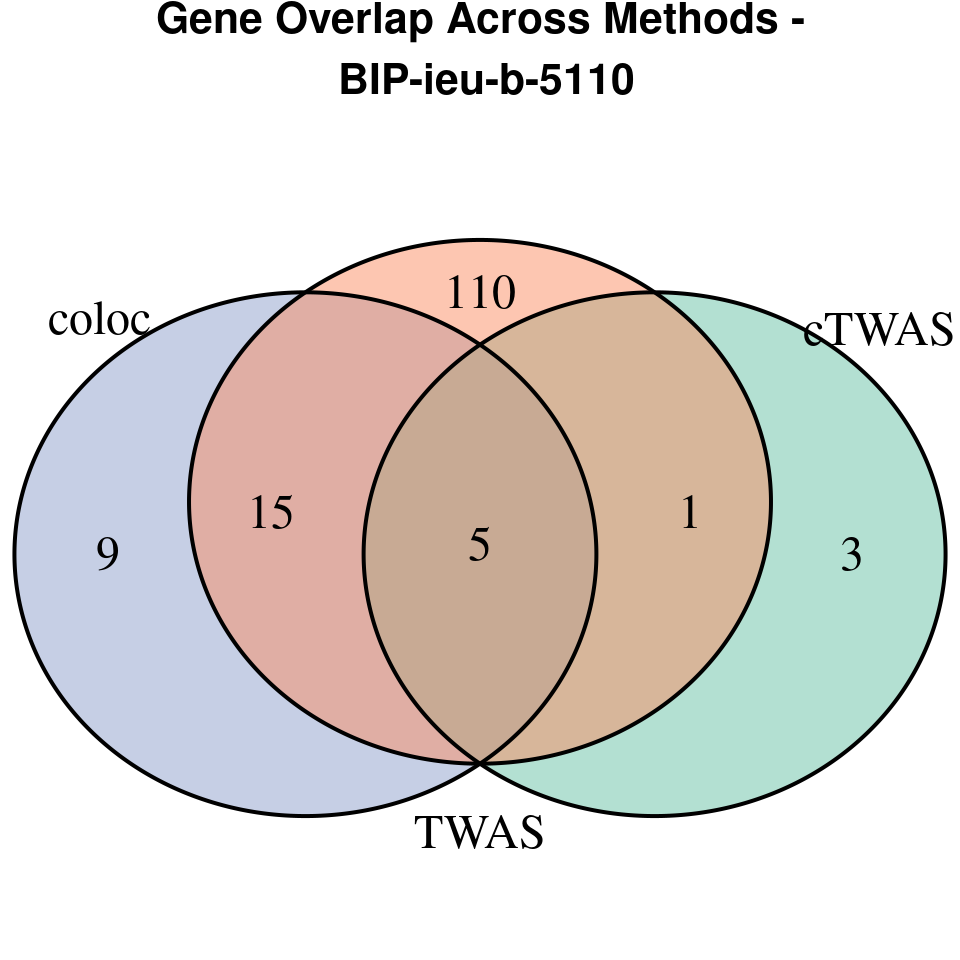
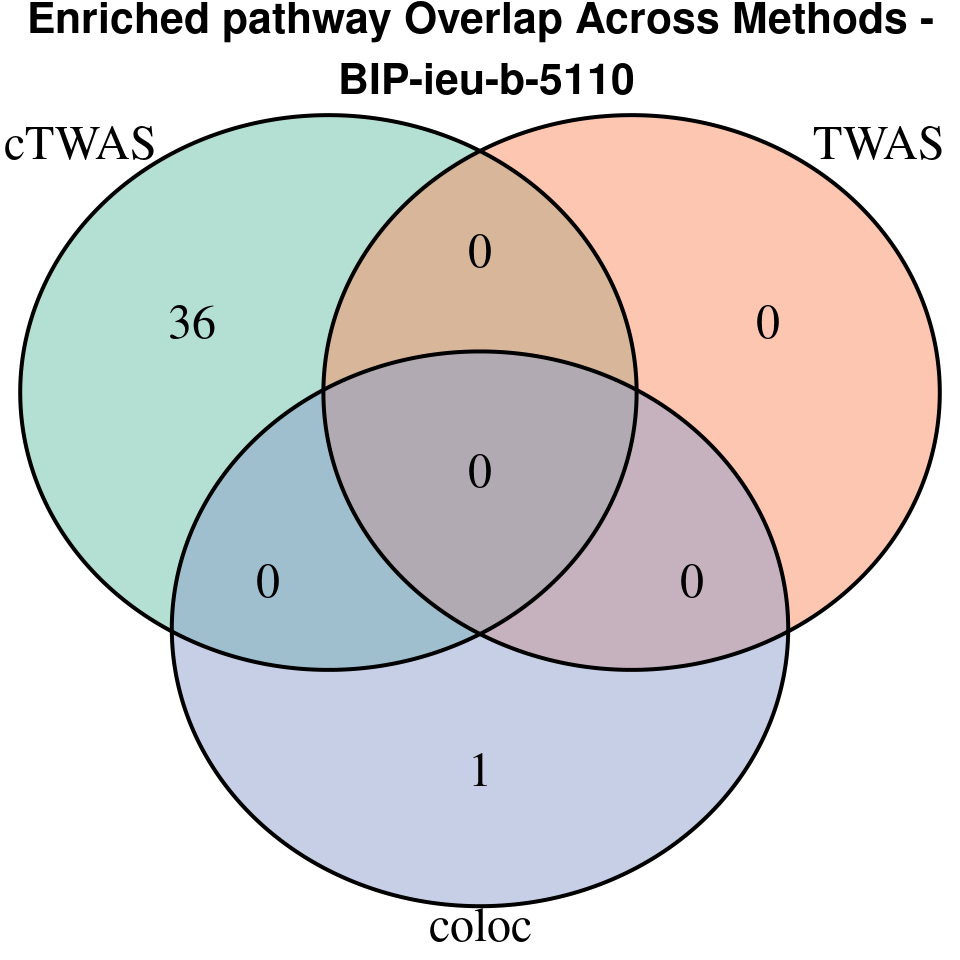 [1]
“PD-ieu-b-7”
[1]
“PD-ieu-b-7”
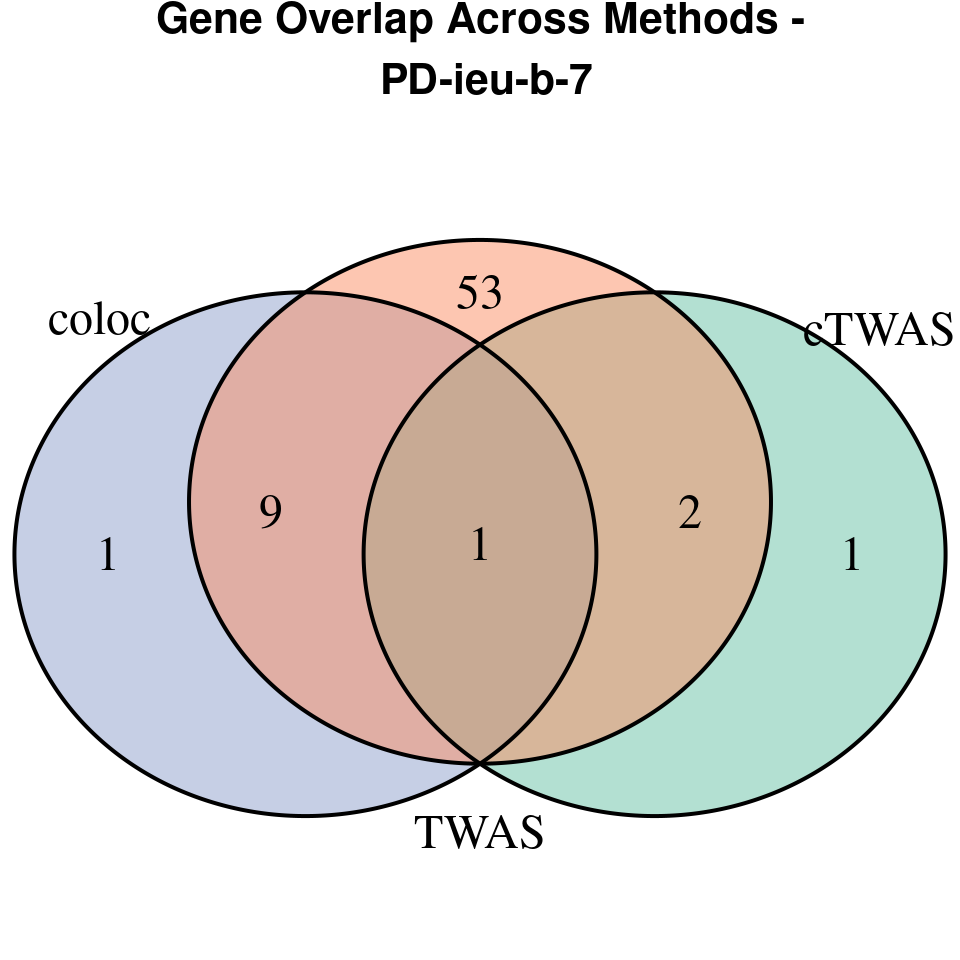
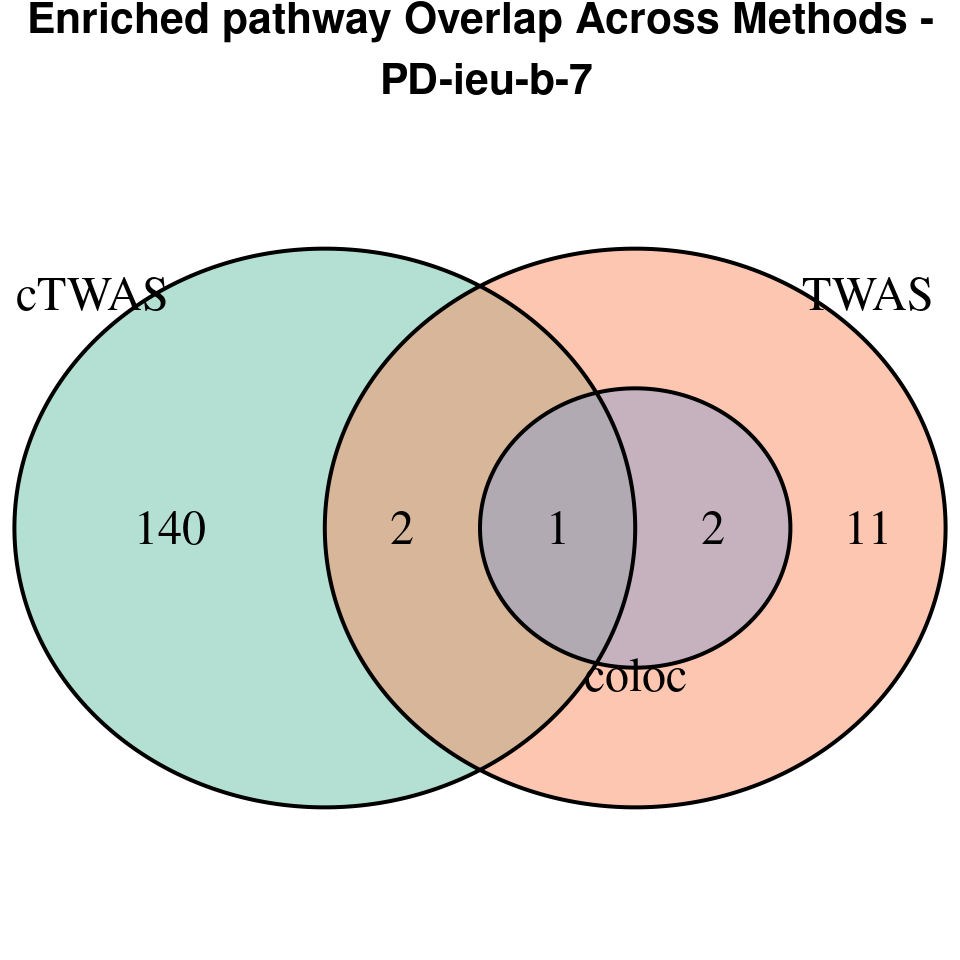 [1]
“NS-ukb-a-230”
[1]
“NS-ukb-a-230”