LDL - Ten tissues
2023-12-3
Last updated: 2023-12-03
Checks: 6 1
Knit directory: multigroup_ctwas_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20231112) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version a69235e. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Unstaged changes:
Modified: analysis/LDL_ten_tissues.Rmd
Modified: analysis/simulation_4tissues_correlated.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/LDL_ten_tissues.Rmd) and HTML (docs/LDL_ten_tissues.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | a69235e | sq-96 | 2023-12-03 | update |
| html | a69235e | sq-96 | 2023-12-03 | update |
| Rmd | 11b528b | sq-96 | 2023-12-03 | update |
| html | 11b528b | sq-96 | 2023-12-03 | update |
| html | 7837083 | sq-96 | 2023-12-03 | update |
| Rmd | e1fd609 | sq-96 | 2023-12-03 | update |
| html | e1fd609 | sq-96 | 2023-12-03 | update |
Load ctwas results
outputdir <- "/project/xinhe/shengqian/cTWAS/cTWAS_analysis/data/LDL_multi_tissue/"
outname <- "LDL_Liver_ctwas"
gwas_n <- 343621
thin <- 0.1
ctwas_parameters <- ctwas:::ctwas_summarize_parameters(outputdir = outputdir,
outname = outname,
gwas_n = 343621,
thin = 0.1)ctwas_parameters$group_size SNP Liver Adipose_Subcutaneous
7405450 9676 11783
Brain_Cerebellum Adipose_Visceral_Omentum Whole_Blood
10862 11586 10028
Lung Artery_Tibial Heart_Left_Ventricle
12012 11728 10340
Stomach Pancreas
10897 10664 ctwas_parameters$group_prior SNP Liver Adipose_Subcutaneous
0.0001256788 0.0059682025 0.0051052884
Brain_Cerebellum Adipose_Visceral_Omentum Whole_Blood
0.0025228508 0.0007715901 0.0010434611
Lung Artery_Tibial Heart_Left_Ventricle
0.0069704792 0.0003173744 0.0035484192
Stomach Pancreas
0.0096415389 0.0104608915 ctwas_parameters$group_prior_var SNP Liver Adipose_Subcutaneous
10.849644 45.616344 5.442887
Brain_Cerebellum Adipose_Visceral_Omentum Whole_Blood
50.884682 11.457110 88.062796
Lung Artery_Tibial Heart_Left_Ventricle
6.723151 894.782000 6.249513
Stomach Pancreas
7.437171 5.007625 ctwas_parameters$enrichment Liver Adipose_Subcutaneous Brain_Cerebellum
47.487727 40.621702 20.073791
Adipose_Visceral_Omentum Whole_Blood Lung
6.139380 8.302600 55.462630
Artery_Tibial Heart_Left_Ventricle Stomach
2.525281 28.234022 76.715689
Pancreas
83.235103 ctwas_parameters$group_pve SNP Liver Adipose_Subcutaneous
0.0293866048 0.0076662008 0.0009528528
Brain_Cerebellum Adipose_Visceral_Omentum Whole_Blood
0.0040579692 0.0002980681 0.0026816579
Lung Artery_Tibial Heart_Left_Ventricle
0.0016382158 0.0096924472 0.0006673013
Stomach Pancreas
0.0022739524 0.0016257019 ctwas_parameters$convergence_plot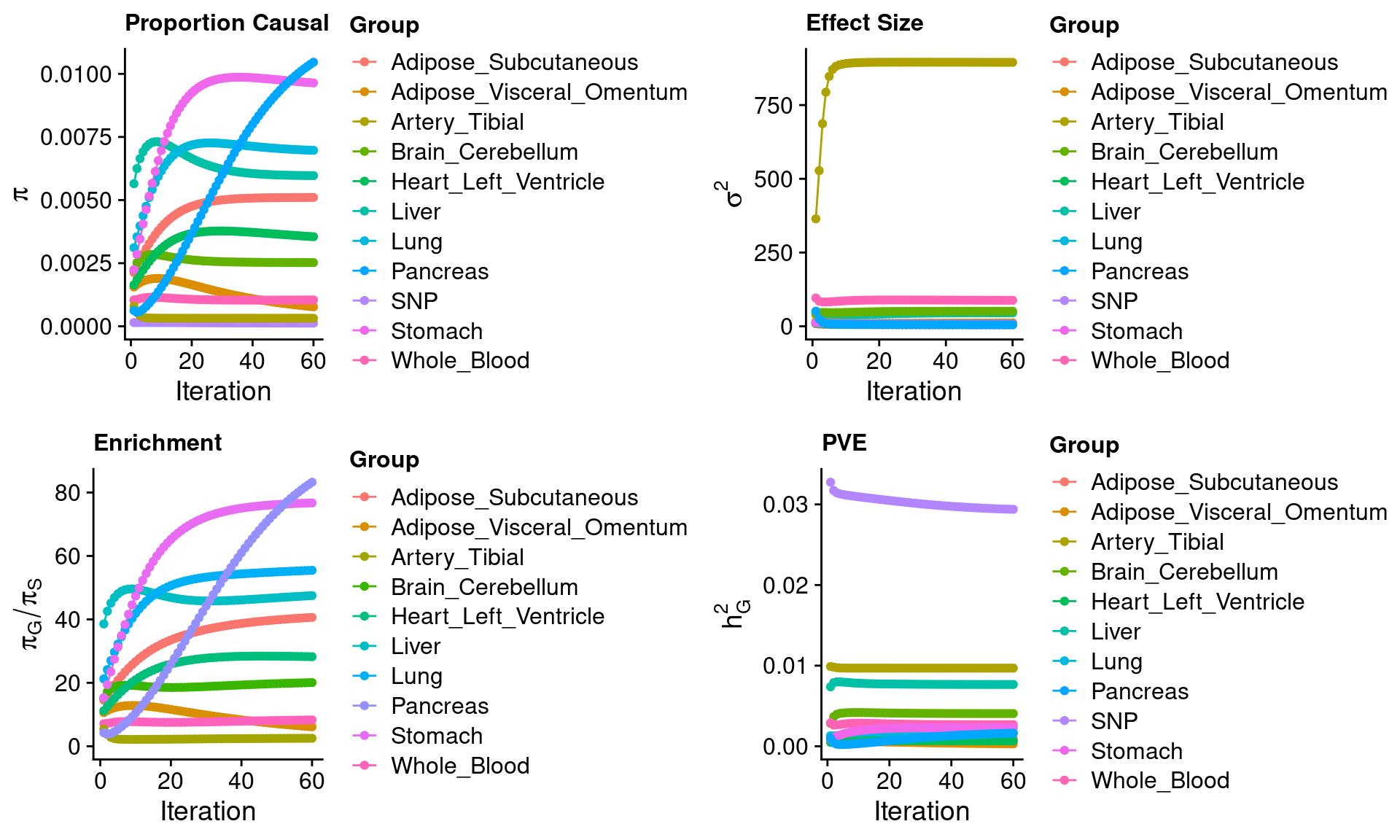
sessionInfo()R version 4.2.0 (2022-04-22)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] cowplot_1.1.1 ggplot2_3.4.4 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] tidyselect_1.2.0 xfun_0.32 bslib_0.4.0 lattice_0.20-45
[5] pgenlibr_0.3.2 ctwas_0.1.40 generics_0.1.3 colorspace_2.0-3
[9] vctrs_0.6.4 htmltools_0.5.3 yaml_2.3.5 utf8_1.2.2
[13] rlang_1.1.1 jquerylib_0.1.4 later_1.3.0 pillar_1.8.1
[17] withr_2.5.0 DBI_1.1.3 glue_1.6.2 foreach_1.5.2
[21] lifecycle_1.0.3 stringr_1.5.0 munsell_0.5.0 gtable_0.3.1
[25] codetools_0.2-18 evaluate_0.16 labeling_0.4.2 knitr_1.40
[29] callr_3.7.2 fastmap_1.1.0 httpuv_1.6.5 ps_1.7.1
[33] fansi_1.0.3 highr_0.9 logging_0.10-108 Rcpp_1.0.9
[37] promises_1.2.0.1 scales_1.2.1 cachem_1.0.6 jsonlite_1.8.0
[41] farver_2.1.1 fs_1.5.2 digest_0.6.29 stringi_1.7.8
[45] processx_3.7.0 dplyr_1.0.10 getPass_0.2-2 rprojroot_2.0.3
[49] grid_4.2.0 cli_3.6.1 tools_4.2.0 magrittr_2.0.3
[53] sass_0.4.2 tibble_3.1.8 whisker_0.4 pkgconfig_2.0.3
[57] Matrix_1.5-3 data.table_1.14.2 assertthat_0.2.1 rmarkdown_2.16
[61] httr_1.4.4 rstudioapi_0.14 iterators_1.0.14 R6_2.5.1
[65] git2r_0.30.1 compiler_4.2.0