Simulation Results using Multiple Tissues
shengqian
2023-09-30
Last updated: 2023-10-05
Checks: 6 1
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 9ed1bc7. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .ipynb_checkpoints/
Untracked files:
Untracked: LDL_LDLR_locus1.pdf
Untracked: LDL_TEME199_genetrack.pdf
Untracked: LDL_TEME199_locus.pdf
Untracked: Proposal plots.R
Untracked: RGS14.pdf
Untracked: RNF186.pdf
Untracked: Rplots.pdf
Untracked: SCZ_annotation.xlsx
Untracked: SLC8B1.pdf
Untracked: analysis/.ipynb_checkpoints/
Untracked: cache/
Untracked: code/.ipynb_checkpoints/
Untracked: data/.ipynb_checkpoints/
Untracked: data/FUMA_output/
Untracked: data/GO_Terms/
Untracked: data/GTEx_Analysis_v8_eQTL.tar
Untracked: data/G_list.RData
Untracked: data/IBD_ME/
Untracked: data/LB/
Untracked: data/LDL/
Untracked: data/LDL_E_S/
Untracked: data/LDL_E_S_M/
Untracked: data/LDL_M/
Untracked: data/LDL_S/
Untracked: data/LungCancer_E_S_M/
Untracked: data/OxHb/
Untracked: data/PGC3_SCZ_wave3_public.v2.tsv
Untracked: data/Predictive_Models/
Untracked: data/ProstateCancer_E_S_M/
Untracked: data/Supplementary Table 15 - MAGMA.xlsx
Untracked: data/Supplementary Table 20 - Prioritised Genes.xlsx
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/WhiteBlood_E/
Untracked: data/WhiteBlood_E_M/
Untracked: data/WhiteBlood_E_S_M/
Untracked: data/WhiteBlood_E_S_M_PC/
Untracked: data/WhiteBlood_M/
Untracked: data/WhiteBlood_M_compare/
Untracked: data/WhiteBlood_M_enet/
Untracked: data/WhiteBlood_M_mashr/
Untracked: data/cpg_annot.RData
Untracked: data/eqtl/
Untracked: data/gencode.v26.GRCh38.genes.gtf
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/gwas_sumstats/
Untracked: data/magma.genes.out
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/mqtl/
Untracked: data/notes.txt
Untracked: data/scz_2018.RDS
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/test/
Untracked: hist.pdf
Untracked: ld_pip.pdf
Untracked: submit.sh
Untracked: temp_LDR/
Untracked: test-B1.snpgwas.txt
Unstaged changes:
Deleted: analysis/Atrial_Fibrillation_Heart_Atrial_Appendage.Rmd
Deleted: analysis/Atrial_Fibrillation_Heart_Left_Ventricle.Rmd
Deleted: analysis/Autism_Brain_Amygdala.Rmd
Deleted: analysis/Autism_Brain_Anterior_cingulate_cortex_BA24.Rmd
Deleted: analysis/Autism_Brain_Caudate_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Cerebellar_Hemisphere.Rmd
Deleted: analysis/Autism_Brain_Cerebellum.Rmd
Deleted: analysis/Autism_Brain_Cortex.Rmd
Deleted: analysis/Autism_Brain_Frontal_Cortex_BA9.Rmd
Deleted: analysis/Autism_Brain_Hippocampus.Rmd
Deleted: analysis/Autism_Brain_Hypothalamus.Rmd
Deleted: analysis/Autism_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Putamen_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Spinal_cord_cervical_c-1.Rmd
Deleted: analysis/Autism_Brain_Substantia_nigra.Rmd
Deleted: analysis/BMI_Brain_Amygdala.Rmd
Deleted: analysis/BMI_Brain_Amygdala_S.Rmd
Deleted: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24.Rmd
Deleted: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24_S.Rmd
Deleted: analysis/BMI_Brain_Caudate_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Caudate_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Cerebellar_Hemisphere.Rmd
Deleted: analysis/BMI_Brain_Cerebellar_Hemisphere_S.Rmd
Deleted: analysis/BMI_Brain_Cerebellum.Rmd
Deleted: analysis/BMI_Brain_Cerebellum_S.Rmd
Deleted: analysis/BMI_Brain_Cortex.Rmd
Deleted: analysis/BMI_Brain_Cortex_S.Rmd
Deleted: analysis/BMI_Brain_Frontal_Cortex_BA9.Rmd
Deleted: analysis/BMI_Brain_Frontal_Cortex_BA9_S.Rmd
Deleted: analysis/BMI_Brain_Hippocampus.Rmd
Deleted: analysis/BMI_Brain_Hippocampus_S.Rmd
Deleted: analysis/BMI_Brain_Hypothalamus.Rmd
Deleted: analysis/BMI_Brain_Hypothalamus_S.Rmd
Deleted: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Putamen_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Putamen_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Spinal_cord_cervical_c-1.Rmd
Deleted: analysis/BMI_Brain_Spinal_cord_cervical_c-1_S.Rmd
Deleted: analysis/BMI_Brain_Substantia_nigra.Rmd
Deleted: analysis/BMI_Brain_Substantia_nigra_S.Rmd
Deleted: analysis/BMI_S_results.Rmd
Deleted: analysis/Glucose_Adipose_Subcutaneous.Rmd
Deleted: analysis/Glucose_Adipose_Visceral_Omentum.Rmd
Modified: analysis/simulation_test.Rmd
Deleted: code/WhiteBlood_M_ener_out/WhiteBlood_WholeBlood.out
Deleted: code/White_Blood_M_out/White_Blood_BreastMammary.err
Deleted: code/White_Blood_M_out/White_Blood_BreastMammary.out
Deleted: code/White_Blood_M_out/White_Blood_ColonTransverse.err
Deleted: code/White_Blood_M_out/White_Blood_ColonTransverse.out
Deleted: code/White_Blood_M_out/White_Blood_KidneyCortex.err
Deleted: code/White_Blood_M_out/White_Blood_KidneyCortex.out
Deleted: code/White_Blood_M_out/White_Blood_Lung.err
Deleted: code/White_Blood_M_out/White_Blood_Lung.out
Deleted: code/White_Blood_M_out/White_Blood_MuscleSkeletal.err
Deleted: code/White_Blood_M_out/White_Blood_MuscleSkeletal.out
Deleted: code/White_Blood_M_out/White_Blood_Ovary.err
Deleted: code/White_Blood_M_out/White_Blood_Ovary.out
Deleted: code/White_Blood_M_out/White_Blood_Prostate.err
Deleted: code/White_Blood_M_out/White_Blood_Prostate.out
Deleted: code/White_Blood_M_out/White_Blood_Testis.err
Deleted: code/White_Blood_M_out/White_Blood_Testis.out
Deleted: code/White_Blood_M_out/White_Blood_WholeBlood.err
Deleted: code/White_Blood_M_out/White_Blood_WholeBlood.out
Deleted: code/run_IBD_ctwas_rss_LDR_ME.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/simulation_test.Rmd) and HTML (docs/simulation_test.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 9ed1bc7 | sq-96 | 2023-10-05 | update |
| html | 9ed1bc7 | sq-96 | 2023-10-05 | update |
Simulation 1: Two causal tissues with equal PVE
Separate effect size parameters
For the cTWAS analysis, each tissue had its own prior inclusion parameter end effect size parameter.
results_dir <- "/project2/xinhe/shengqian/cTWAS/cTWAS_simulation/simulation_test_merge/"
runtag = "ukb-s80.45-liv_wb"
configtag <- 2
simutags <- paste(1, 1:5, sep = "-")
thin <- 0.1
sample_size <- 45000
PIP_threshold <- 0.8
results_df <- data.frame(simutag=as.character(),
n_causal=as.integer(),
n_causal_combined=as.integer(),
n_detected_pip=as.integer(),
n_detected_pip_in_causal=as.integer(),
n_detected_comb_pip=as.integer(),
n_detected_comb_pip_in_causal=as.integer(),
pve_snp=as.numeric(),
pve_weight1=as.numeric(),
pve_weight2=as.numeric(),
pve_weight3=as.numeric(),
prior_weight1=as.numeric(),
prior_weight2=as.numeric(),
prior_weight3=as.numeric(),
prior_var_snp=as.numeric(),
prior_var_weight1=as.numeric(),
prior_var_weight2=as.numeric(),
prior_var_weight3=as.numeric())
for (i in 1:length(simutags)){
simutag <- simutags[i]
#load genes with true simulated effect
load(paste0(results_dir, runtag, "_simu", simutag, "-pheno.Rd"))
true_genes <- unlist(sapply(1:22, function(x){phenores$batch[[x]]$id.cgene}))
true_genes_combined <- unique(sapply(true_genes, function(x){unlist(strsplit(x, "[|]"))[1]}))
#load cTWAS results
ctwas_res <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR.susieIrss.txt"))
ctwas_gene_res <- ctwas_res[ctwas_res$type!="SNP",]
#number of causal genes
n_causal <- length(true_genes)
n_causal_combined <- length(true_genes_combined)
#number of gene+tissue combinations with cTWAS PIP > threshold
n_ctwas_genes <- sum(ctwas_gene_res$susie_pip > PIP_threshold)
#number of cTWAS genes that are causal
n_causal_detected <- sum(ctwas_gene_res$id[ctwas_gene_res$susie_pip > PIP_threshold] %in% true_genes)
#collapse gene+tissues to genes and compute combined PIP
ctwas_gene_res$gene <- sapply(ctwas_gene_res$id, function(x){unlist(strsplit(x,"[|]"))[1]})
ctwas_gene_res_combined <- aggregate(ctwas_gene_res$susie_pip, by=list(ctwas_gene_res$gene), FUN=sum)
colnames(ctwas_gene_res_combined) <- c("gene", "pip_combined")
#number of genes with combined PIP > threshold
n_ctwas_genes_combined <- sum(ctwas_gene_res_combined$pip_combined > PIP_threshold)
#number of cTWAS genes using combined PIP that are causal
n_causal_detected_combined <- sum(ctwas_gene_res_combined$gene[ctwas_gene_res_combined$pip_combined > PIP_threshold] %in% true_genes_combined)
#collect number of SNPs analyzed by cTWAS
ctwas_res_s1 <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR.s1.susieIrss.txt"))
n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
rm(ctwas_res_s1)
#load estimated parameters
load(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR.s2.susieIrssres.Rd"))
#estimated group prior (all iterations)
estimated_group_prior_all <- group_prior_rec
estimated_group_prior_all["SNP",] <- estimated_group_prior_all["SNP",]*thin #adjust parameter to account for thin argument
#estimated group prior variance (all iterations)
estimated_group_prior_var_all <- group_prior_var_rec
#set group size
group_size <- c(table(ctwas_gene_res$type), structure(n_snps, names="SNP"))
group_size <- group_size[rownames(estimated_group_prior_all)]
#estimated group PVE (all iterations)
estimated_group_pve_all <- estimated_group_prior_var_all*estimated_group_prior_all*group_size/sample_size #check PVE calculation
results_current <- data.frame(simutag=as.character(simutag),
n_causal=as.integer(n_causal),
n_causal_combined=as.integer(n_causal_combined),
n_detected_pip=as.integer(n_ctwas_genes),
n_detected_pip_in_causal=as.integer(n_causal_detected),
n_detected_comb_pip=as.integer(n_ctwas_genes_combined),
n_detected_comb_pip_in_causal=as.integer(n_causal_detected_combined),
pve_snp=as.numeric(rev(estimated_group_pve_all["SNP",])[1]),
pve_weight1=as.numeric(rev(estimated_group_pve_all["Liver",])[1]),
pve_weight2=as.numeric(rev(estimated_group_pve_all["Whole_Blood",])[1]),
pve_weight3=as.numeric(rev(estimated_group_pve_all["Brain_Cerebellum",])[1]),
prior_snp=as.numeric(rev(estimated_group_prior_var_all["SNP",])[1]),
prior_weight1=as.numeric(rev(estimated_group_prior_all["Liver",])[1]),
prior_weight2=as.numeric(rev(estimated_group_prior_all["Whole_Blood",])[1]),
prior_weight3=as.numeric(rev(estimated_group_prior_all["Brain_Cerebellum",])[1]),
prior_var_snp=as.numeric(rev(estimated_group_prior_var_all["SNP",])[1]),
prior_var_weight1=as.numeric(rev(estimated_group_prior_var_all["Liver",])[1]),
prior_var_weight2=as.numeric(rev(estimated_group_prior_var_all["Whole_Blood",])[1]),
prior_var_weight3=as.numeric(rev(estimated_group_prior_var_all["Brain_Cerebellum",])[1]))
results_df <- rbind(results_df, results_current)
}
#results using PIP threshold (gene+tissue)
results_df[,c("simutag", "n_causal", "n_detected_pip", "n_detected_pip_in_causal")] simutag n_causal n_detected_pip n_detected_pip_in_causal
1 1-1 215 67 53
2 1-2 250 91 74
3 1-3 216 67 48
4 1-4 231 62 45
5 1-5 232 66 46#mean percent causal using PIP > 0.8
sum(results_df$n_detected_pip_in_causal)/sum(results_df$n_detected_pip)[1] 0.7535411#results using combined PIP threshold
results_df[,c("simutag", "n_causal_combined", "n_detected_comb_pip", "n_detected_comb_pip_in_causal")] simutag n_causal_combined n_detected_comb_pip n_detected_comb_pip_in_causal
1 1-1 214 90 68
2 1-2 247 114 92
3 1-3 216 97 67
4 1-4 231 87 63
5 1-5 230 85 59#mean percent causal using combined PIP > 0.8
sum(results_df$n_detected_comb_pip_in_causal)/sum(results_df$n_detected_comb_pip)[1] 0.7378436#prior inclusion and mean prior inclusion
results_df[,c(which(colnames(results_df)=="simutag"), setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df))))] simutag prior_snp prior_weight1 prior_weight2 prior_weight3
1 1-1 13.98029 0.01049760 0.010440150 0.001554739
2 1-2 14.32527 0.01562886 0.030946154 0.003375131
3 1-3 13.24313 0.01599995 0.011503706 0.001654758
4 1-4 12.93168 0.01632716 0.006898015 0.001867553
5 1-5 14.78867 0.01116179 0.011345179 0.004362640colMeans(results_df[,setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df)))]) prior_snp prior_weight1 prior_weight2 prior_weight3
13.853807579 0.013923071 0.014226641 0.002562964 #prior variance and mean prior variance
results_df[,c(which(colnames(results_df)=="simutag"), grep("prior_var", names(results_df)))] simutag prior_var_snp prior_var_weight1 prior_var_weight2 prior_var_weight3
1 1-1 13.98029 24.70155 14.25856 40.345898
2 1-2 14.32527 22.03369 10.92709 2.446965
3 1-3 13.24313 12.81897 14.70263 13.803416
4 1-4 12.93168 16.48358 44.99221 4.299581
5 1-5 14.78867 27.58522 23.39390 15.000354colMeans(results_df[,grep("prior_var", names(results_df))]) prior_var_snp prior_var_weight1 prior_var_weight2 prior_var_weight3
13.85381 20.72460 21.65488 15.17924 #PVE and mean PVE
results_df[,c(which(colnames(results_df)=="simutag"), grep("pve", names(results_df)))] simutag pve_snp pve_weight1 pve_weight2 pve_weight3
1 1-1 0.3839227 0.04261279 0.02702663 0.012263889
2 1-2 0.3668868 0.05659007 0.06139325 0.001614692
3 1-3 0.4037207 0.03370524 0.03070735 0.004465732
4 1-4 0.4050848 0.04422702 0.05634702 0.001569894
5 1-5 0.4118573 0.05059830 0.04818629 0.012794470colMeans(results_df[,grep("pve", names(results_df))]) pve_snp pve_weight1 pve_weight2 pve_weight3
0.394294484 0.045546683 0.044732107 0.006541735 #store results for figure
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS G - Ind Var",
count=results_df$n_detected_comb_pip-results_df$n_detected_comb_pip_in_causal,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS G - Ind Var",
count=results_df$n_detected_comb_pip_in_causal,
ifcausal=T))Individual tissue analyses
For the cTWAS analysis, each tissue was analyzed individually and the results were combined.
results_dir <- "/project2/xinhe/shengqian/cTWAS/cTWAS_simulation/simulation_test_merge/"
runtag = "ukb-s80.45-liv_wb"
configtag <- 2
simutags <- paste(1, 1:5, sep = "-")
thin <- 0.1
sample_size <- 45000
PIP_threshold <- 0.8
results_df <- data.frame(simutag=as.character(),
n_causal_combined=as.integer(),
n_detected_weight1=as.integer(),
n_detected_in_causal_weight1=as.integer(),
n_detected_weight2=as.integer(),
n_detected_in_causal_weight2=as.integer(),
n_detected_weight3=as.integer(),
n_detected_in_causal_weight3=as.integer(),
n_detected_combined=as.integer(),
n_detected_in_causal_combined=as.integer())
for (i in 1:length(simutags)){
simutag <- simutags[i]
#load genes with true simulated effect
load(paste0(results_dir, runtag, "_simu", simutag, "-pheno.Rd"))
true_genes <- unlist(sapply(1:22, function(x){phenores$batch[[x]]$id.cgene}))
true_genes_combined <- unique(sapply(true_genes, function(x){unlist(strsplit(x, "[|]"))[1]}))
#number of causal genes
n_causal_combined <- length(true_genes_combined)
#load cTWAS results for weight1
ctwas_res <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR_weight1.susieIrss.txt"))
ctwas_gene_res <- ctwas_res[ctwas_res$type!="SNP",]
#number of genes with cTWAS PIP > threshold in weight1
ctwas_genes_weight1 <- ctwas_gene_res$id[ctwas_gene_res$susie_pip > PIP_threshold]
n_ctwas_genes_weight1 <- length(ctwas_genes_weight1)
n_causal_detected_weight1 <- sum(ctwas_genes_weight1 %in% true_genes_combined)
#load cTWAS results for weight2
ctwas_res <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR_weight2.susieIrss.txt"))
ctwas_gene_res <- ctwas_res[ctwas_res$type!="SNP",]
#number of genes with cTWAS PIP > threshold in weight1
ctwas_genes_weight2 <- ctwas_gene_res$id[ctwas_gene_res$susie_pip > PIP_threshold]
n_ctwas_genes_weight2 <- length(ctwas_genes_weight2)
n_causal_detected_weight2 <- sum(ctwas_genes_weight2 %in% true_genes_combined)
#load cTWAS results for weight3
ctwas_res <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR_weight3.susieIrss.txt"))
ctwas_gene_res <- ctwas_res[ctwas_res$type!="SNP",]
#number of genes with cTWAS PIP > threshold in weight3
ctwas_genes_weight3 <- ctwas_gene_res$id[ctwas_gene_res$susie_pip > PIP_threshold]
n_ctwas_genes_weight3 <- length(ctwas_genes_weight3)
n_causal_detected_weight3 <- sum(ctwas_genes_weight3 %in% true_genes_combined)
#combined analysis
ctwas_genes_combined <- unique(c(ctwas_genes_weight1, ctwas_genes_weight2, ctwas_genes_weight3))
n_ctwas_genes_combined <- length(ctwas_genes_combined)
n_causal_detected_combined <- sum(ctwas_genes_combined %in% true_genes_combined)
results_current <- data.frame(simutag=as.character(simutag),
n_causal_combined=as.integer(n_causal_combined),
n_detected_weight1=as.integer(n_ctwas_genes_weight1),
n_detected_in_causal_weight1=as.integer(n_causal_detected_weight1),
n_detected_weight2=as.integer(n_ctwas_genes_weight2),
n_detected_in_causal_weight2=as.integer(n_causal_detected_weight2),
n_detected_weight3=as.integer(n_ctwas_genes_weight3),
n_detected_in_causal_weight3=as.integer(n_causal_detected_weight3),
n_detected_combined=as.integer(n_ctwas_genes_combined),
n_detected_in_causal_combined=as.integer(n_causal_detected_combined))
results_df <- rbind(results_df, results_current)
}
#results using weight1
results_df[,c("simutag", colnames(results_df)[grep("weight1", colnames(results_df))])] simutag n_detected_weight1 n_detected_in_causal_weight1
1 1-1 60 47
2 1-2 79 61
3 1-3 61 42
4 1-4 54 38
5 1-5 55 42#mean percent causal using PIP > 0.8 for weight1
sum(results_df$n_detected_in_causal_weight1)/sum(results_df$n_detected_weight1)[1] 0.7443366#results using weight2
results_df[,c("simutag", colnames(results_df)[grep("weight2", colnames(results_df))])] simutag n_detected_weight2 n_detected_in_causal_weight2
1 1-1 52 33
2 1-2 81 59
3 1-3 64 45
4 1-4 55 39
5 1-5 50 33#mean percent causal using PIP > 0.8 for weight1
sum(results_df$n_detected_in_causal_weight2)/sum(results_df$n_detected_weight2)[1] 0.692053#results using weight3
results_df[,c("simutag", colnames(results_df)[grep("weight3", colnames(results_df))])] simutag n_detected_weight3 n_detected_in_causal_weight3
1 1-1 28 13
2 1-2 34 17
3 1-3 27 12
4 1-4 26 15
5 1-5 32 14#mean percent causal using PIP > 0.8 for weight3
sum(results_df$n_detected_in_causal_weight3)/sum(results_df$n_detected_weight3)[1] 0.4829932#results using combined analysis
results_df[,c("simutag", colnames(results_df)[grep("combined", colnames(results_df))])] simutag n_causal_combined n_detected_combined n_detected_in_causal_combined
1 1-1 214 107 71
2 1-2 247 151 101
3 1-3 216 113 68
4 1-4 231 104 66
5 1-5 230 102 62#mean percent causal using PIP > 0.8 for weight3
sum(results_df$n_detected_in_causal_combined)/sum(results_df$n_detected_combined)[1] 0.6377816#store results for figure
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight1",
count=results_df$n_detected_weight1-results_df$n_detected_in_causal_weight1,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight1",
count=results_df$n_detected_in_causal_weight1,
ifcausal=T))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight2",
count=results_df$n_detected_weight2-results_df$n_detected_in_causal_weight2,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight2",
count=results_df$n_detected_in_causal_weight2,
ifcausal=T))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight3",
count=results_df$n_detected_weight3-results_df$n_detected_in_causal_weight3,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight3",
count=results_df$n_detected_in_causal_weight3,
ifcausal=T))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Union",
count=results_df$n_detected_combined-results_df$n_detected_in_causal_combined,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Union",
count=results_df$n_detected_in_causal_combined,
ifcausal=T))Figure
For the cTWAS analysis, each tissue was analyzed individually and the results were combined.
plot_df$method <- factor(plot_df$method, levels=c("TWAS", "cTWAS G+T", "cTWAS G", "cTWAS G - Ind Var", "cTWAS Union", "cTWAS Weight1", "cTWAS Weight2", "cTWAS Weight3"))
library(ggpubr)Loading required package: ggplot2colset = c("#ebebeb", "#fb8072")
ggbarplot(plot_df,
x = "method",
y = "count",
add = "mean_se",
fill = "ifcausal",
legend = "none",
ylab="Count",
xlab="",
palette = colset) + grids(linetype = "dashed") + theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1))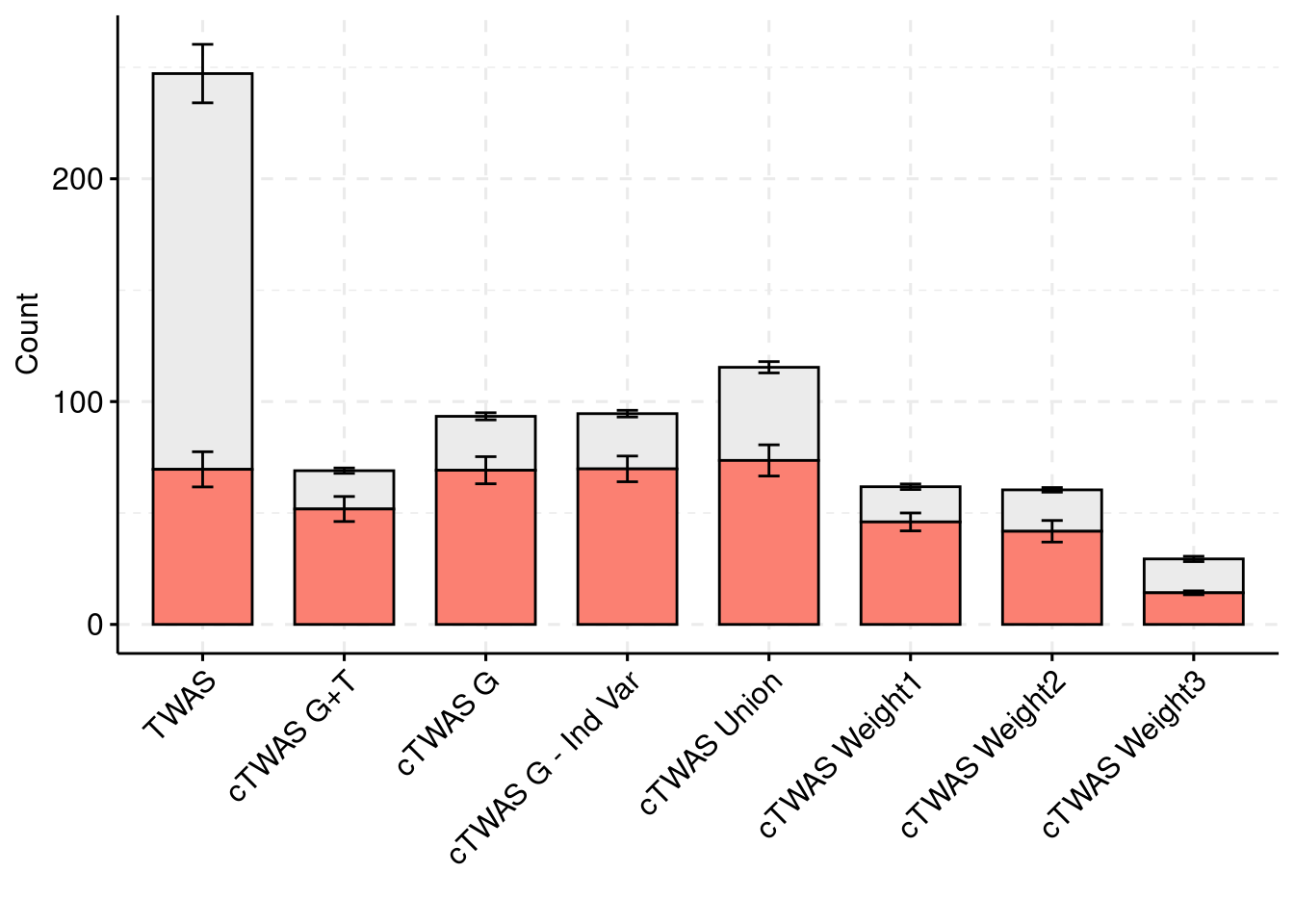
| Version | Author | Date |
|---|---|---|
| 9ed1bc7 | sq-96 | 2023-10-05 |
plot_df_2 <- plot_df
plot_df_2$method %in% c("TWAS", "cTWAS Union", "cTWAS Weight1", "cTWAS Weight2", "cTWAS Weight3") [1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[13] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE
[25] TRUE TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
[37] FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[49] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[61] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[73] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUEplot_df_2$ifcausal <- as.character(plot_df_2$ifcausal+1)
plot_df_2$count[plot_df_2$method=="cTWAS G" & plot_df_2$ifcausal=="2"] <- plot_df_2$count[plot_df_2$method=="cTWAS G" & plot_df_2$ifcausal=="2"] - plot_df_2$count[plot_df_2$method=="cTWAS G+T" & plot_df_2$ifcausal=="2"]
plot_df_2 <- rbind(plot_df_2, data.frame(simutag=simutags,
method="cTWAS G",
count=plot_df_2$count[plot_df_2$method=="cTWAS G+T" & plot_df_2$ifcausal=="2"],
ifcausal="3"))
plot_df_3 <- plot_df_2
plot_df_2 <- plot_df_2[plot_df_2$method %in% c("TWAS", "cTWAS Union", "cTWAS Weight1", "cTWAS Weight2", "cTWAS Weight3"),]
plot_df_2$method <- droplevels(plot_df_2$method)
levels(plot_df_2$method) <- c("TWAS", "cTWAS Union", "cTWAS Primary", "cTWAS Secondary", "cTWAS Null")
plot_df_2$ifcausal[plot_df_2$ifcausal=="1"] <- "FP"
plot_df_2$ifcausal[plot_df_2$ifcausal=="2"] <- "TP"
colset = c("#ebebeb", "#fb8072", "#8dd3c7")
ggbarplot(plot_df_2,
x = "method",
y = "count",
add = "mean_se",
fill = "ifcausal",
legend = "right",
ylab="Count",
xlab="",
palette = colset) + grids(linetype = "dashed") + theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1)) + guides(fill=guide_legend(title=""))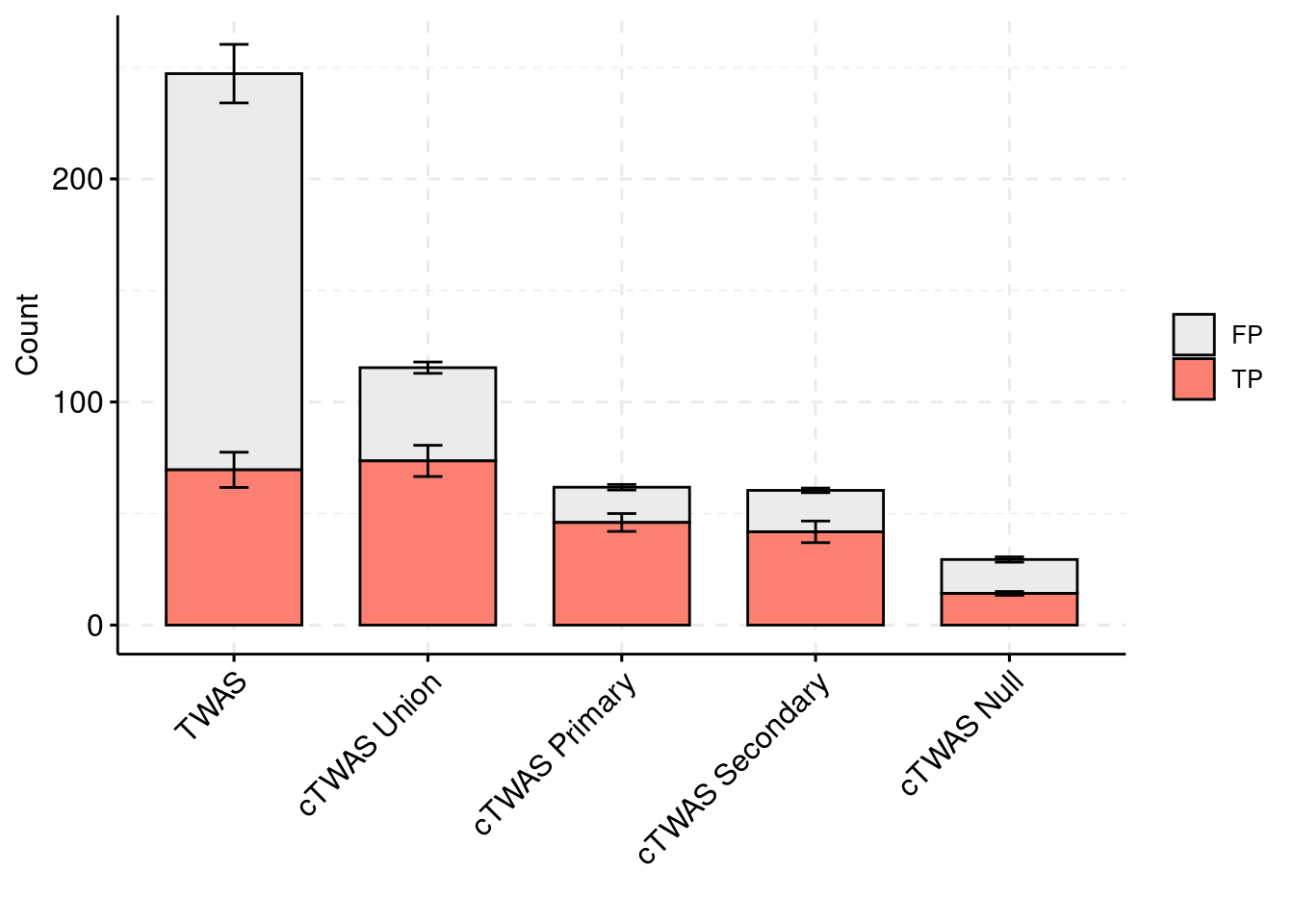
| Version | Author | Date |
|---|---|---|
| 9ed1bc7 | sq-96 | 2023-10-05 |
plot_df_3 <- plot_df_3[plot_df_3$method %in% c("cTWAS G", "cTWAS Union"),]
plot_df_3$method <- droplevels(plot_df_3$method)
levels(plot_df_3$method) <- c("cTWAS Joint", "cTWAS Union")
plot_df_3$method <- relevel(plot_df_3$method, "cTWAS Union")
plot_df_3$ifcausal[plot_df_3$ifcausal=="1"] <- "FP"
plot_df_3$ifcausal[plot_df_3$ifcausal=="2"] <- "TP"
plot_df_3$ifcausal[plot_df_3$ifcausal=="3"] <- "TP and Tissue Identified\nby Joint Analysis"
ggbarplot(plot_df_3,
x = "method",
y = "count",
add = "mean_se",
fill = "ifcausal",
legend = "right",
ylab="Count",
xlab="",
palette = colset) + grids(linetype = "dashed") + theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1)) + guides(fill=guide_legend(title=""))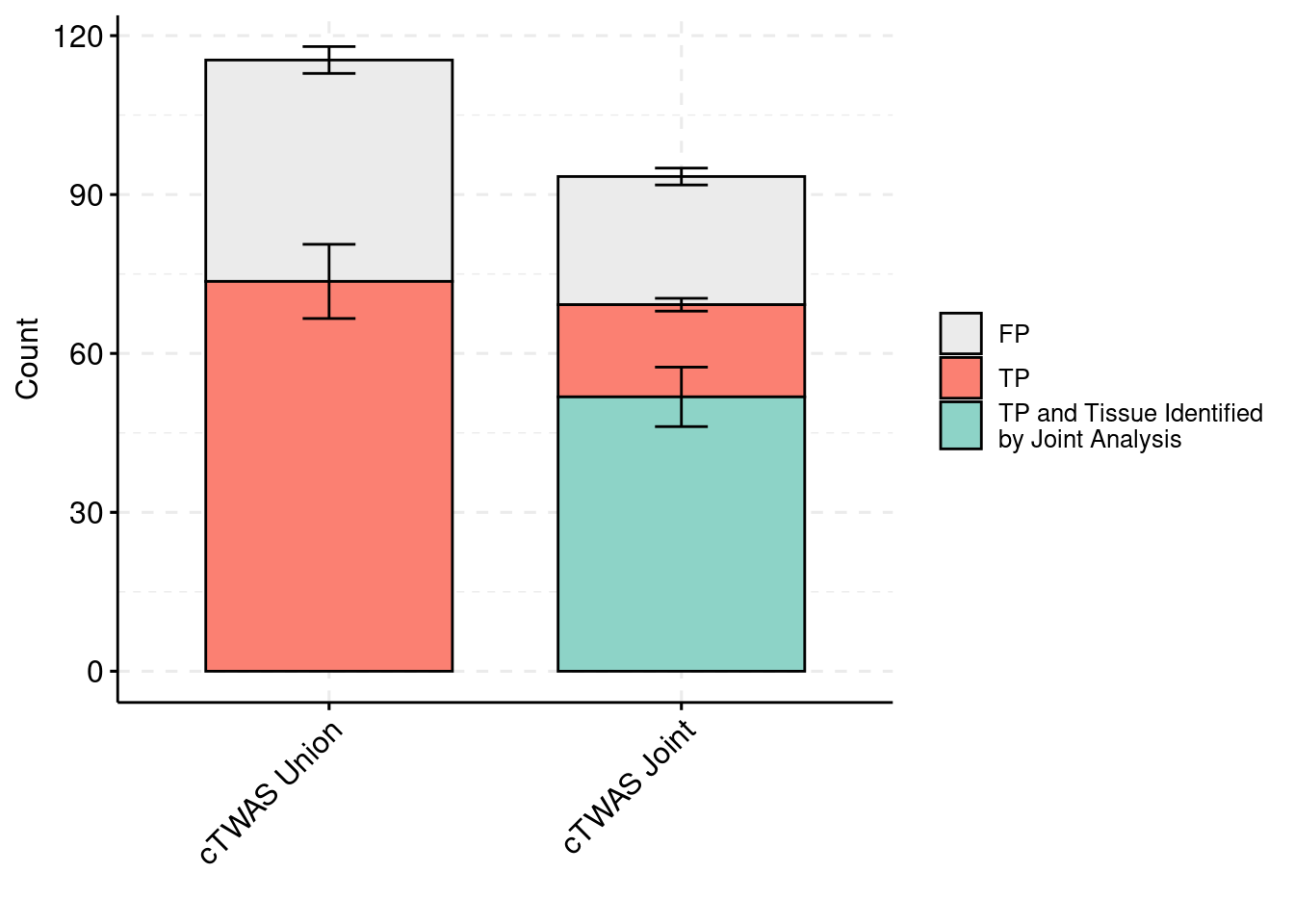
| Version | Author | Date |
|---|---|---|
| 9ed1bc7 | sq-96 | 2023-10-05 |
Simulation 2: Two causal tissues with unequal PVE
Separate effect size parameters
For the cTWAS analysis, each tissue had its own prior inclusion parameter and effect size parameter.
results_dir <- "/project2/xinhe/shengqian/cTWAS/cTWAS_simulation/simulation_test_merge/"
runtag = "ukb-s80.45-liv_wb"
configtag <- 2
simutags <- paste(2, 1:5, sep = "-")
thin <- 0.1
sample_size <- 45000
PIP_threshold <- 0.8
results_df <- data.frame(simutag=as.character(),
n_causal=as.integer(),
n_causal_combined=as.integer(),
n_detected_pip=as.integer(),
n_detected_pip_in_causal=as.integer(),
n_detected_comb_pip=as.integer(),
n_detected_comb_pip_in_causal=as.integer(),
pve_snp=as.numeric(),
pve_weight1=as.numeric(),
pve_weight2=as.numeric(),
pve_weight3=as.numeric(),
prior_weight1=as.numeric(),
prior_weight2=as.numeric(),
prior_weight3=as.numeric(),
prior_var_snp=as.numeric(),
prior_var_weight1=as.numeric(),
prior_var_weight2=as.numeric(),
prior_var_weight3=as.numeric())
for (i in 1:length(simutags)){
simutag <- simutags[i]
#load genes with true simulated effect
load(paste0(results_dir, runtag, "_simu", simutag, "-pheno.Rd"))
true_genes <- unlist(sapply(1:22, function(x){phenores$batch[[x]]$id.cgene}))
true_genes_combined <- unique(sapply(true_genes, function(x){unlist(strsplit(x, "[|]"))[1]}))
#load cTWAS results
ctwas_res <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR.susieIrss.txt"))
ctwas_gene_res <- ctwas_res[ctwas_res$type!="SNP",]
#number of causal genes
n_causal <- length(true_genes)
n_causal_combined <- length(true_genes_combined)
#number of gene+tissue combinations with cTWAS PIP > threshold
n_ctwas_genes <- sum(ctwas_gene_res$susie_pip > PIP_threshold)
#number of cTWAS genes that are causal
n_causal_detected <- sum(ctwas_gene_res$id[ctwas_gene_res$susie_pip > PIP_threshold] %in% true_genes)
#collapse gene+tissues to genes and compute combined PIP
ctwas_gene_res$gene <- sapply(ctwas_gene_res$id, function(x){unlist(strsplit(x,"[|]"))[1]})
ctwas_gene_res_combined <- aggregate(ctwas_gene_res$susie_pip, by=list(ctwas_gene_res$gene), FUN=sum)
colnames(ctwas_gene_res_combined) <- c("gene", "pip_combined")
#number of genes with combined PIP > threshold
n_ctwas_genes_combined <- sum(ctwas_gene_res_combined$pip_combined > PIP_threshold)
#number of cTWAS genes using combined PIP that are causal
n_causal_detected_combined <- sum(ctwas_gene_res_combined$gene[ctwas_gene_res_combined$pip_combined > PIP_threshold] %in% true_genes_combined)
#collect number of SNPs analyzed by cTWAS
ctwas_res_s1 <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR.s1.susieIrss.txt"))
n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
rm(ctwas_res_s1)
#load estimated parameters
load(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR.s2.susieIrssres.Rd"))
#estimated group prior (all iterations)
estimated_group_prior_all <- group_prior_rec
estimated_group_prior_all["SNP",] <- estimated_group_prior_all["SNP",]*thin #adjust parameter to account for thin argument
#estimated group prior variance (all iterations)
estimated_group_prior_var_all <- group_prior_var_rec
#set group size
group_size <- c(table(ctwas_gene_res$type), structure(n_snps, names="SNP"))
group_size <- group_size[rownames(estimated_group_prior_all)]
#estimated group PVE (all iterations)
estimated_group_pve_all <- estimated_group_prior_var_all*estimated_group_prior_all*group_size/sample_size #check PVE calculation
results_current <- data.frame(simutag=as.character(simutag),
n_causal=as.integer(n_causal),
n_causal_combined=as.integer(n_causal_combined),
n_detected_pip=as.integer(n_ctwas_genes),
n_detected_pip_in_causal=as.integer(n_causal_detected),
n_detected_comb_pip=as.integer(n_ctwas_genes_combined),
n_detected_comb_pip_in_causal=as.integer(n_causal_detected_combined),
pve_snp=as.numeric(rev(estimated_group_pve_all["SNP",])[1]),
pve_weight1=as.numeric(rev(estimated_group_pve_all["Liver",])[1]),
pve_weight2=as.numeric(rev(estimated_group_pve_all["Whole_Blood",])[1]),
pve_weight3=as.numeric(rev(estimated_group_pve_all["Brain_Cerebellum",])[1]),
prior_snp=as.numeric(rev(estimated_group_prior_var_all["SNP",])[1]),
prior_weight1=as.numeric(rev(estimated_group_prior_all["Liver",])[1]),
prior_weight2=as.numeric(rev(estimated_group_prior_all["Whole_Blood",])[1]),
prior_weight3=as.numeric(rev(estimated_group_prior_all["Brain_Cerebellum",])[1]),
prior_var_snp=as.numeric(rev(estimated_group_prior_var_all["SNP",])[1]),
prior_var_weight1=as.numeric(rev(estimated_group_prior_var_all["Liver",])[1]),
prior_var_weight2=as.numeric(rev(estimated_group_prior_var_all["Whole_Blood",])[1]),
prior_var_weight3=as.numeric(rev(estimated_group_prior_var_all["Brain_Cerebellum",])[1]))
results_df <- rbind(results_df, results_current)
}
#results using PIP threshold (gene+tissue)
results_df[,c("simutag", "n_causal", "n_detected_pip", "n_detected_pip_in_causal")] simutag n_causal n_detected_pip n_detected_pip_in_causal
1 2-1 215 48 38
2 2-2 250 73 55
3 2-3 216 46 27
4 2-4 231 46 30
5 2-5 232 50 32#mean percent causal using PIP > 0.8
sum(results_df$n_detected_pip_in_causal)/sum(results_df$n_detected_pip)[1] 0.6920152#results using combined PIP threshold
results_df[,c("simutag", "n_causal_combined", "n_detected_comb_pip", "n_detected_comb_pip_in_causal")] simutag n_causal_combined n_detected_comb_pip n_detected_comb_pip_in_causal
1 2-1 214 68 48
2 2-2 247 91 68
3 2-3 216 63 38
4 2-4 231 64 45
5 2-5 230 68 42#mean percent causal using combined PIP > 0.8
sum(results_df$n_detected_comb_pip_in_causal)/sum(results_df$n_detected_comb_pip)[1] 0.680791#prior inclusion and mean prior inclusion
results_df[,c(which(colnames(results_df)=="simutag"), setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df))))] simutag prior_snp prior_weight1 prior_weight2 prior_weight3
1 2-1 14.01892 0.01155677 0.003544356 0.001712023
2 2-2 15.39150 0.01602250 0.029919898 0.003678954
3 2-3 13.17307 0.01706519 0.008016523 0.004806268
4 2-4 12.77783 0.01534327 0.004428347 0.002359614
5 2-5 14.68214 0.01146830 0.005685229 0.006944209colMeans(results_df[,setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df)))]) prior_snp prior_weight1 prior_weight2 prior_weight3
14.008693050 0.014291206 0.010318871 0.003900214 #prior variance and mean prior variance
results_df[,c(which(colnames(results_df)=="simutag"), grep("prior_var", names(results_df)))] simutag prior_var_snp prior_var_weight1 prior_var_weight2 prior_var_weight3
1 2-1 14.01892 22.91043 12.549905 35.390725
2 2-2 15.39150 20.72263 4.463574 3.079414
3 2-3 13.17307 13.49436 4.000902 7.377954
4 2-4 12.77783 17.73800 48.122109 3.573693
5 2-5 14.68214 28.80506 22.130566 8.966683colMeans(results_df[,grep("prior_var", names(results_df))]) prior_var_snp prior_var_weight1 prior_var_weight2 prior_var_weight3
14.00869 20.73410 18.25341 11.67769 #PVE and mean PVE
results_df[,c(which(colnames(results_df)=="simutag"), grep("pve", names(results_df)))] simutag pve_snp pve_weight1 pve_weight2 pve_weight3
1 2-1 0.3861511 0.04351062 0.008075834 0.011845968
2 2-2 0.3811809 0.05456331 0.024246684 0.002214949
3 2-3 0.4025341 0.03784332 0.005823090 0.006932907
4 2-4 0.4041817 0.04472479 0.038689741 0.001648655
5 2-5 0.4129643 0.05428671 0.022842838 0.012173797colMeans(results_df[,grep("pve", names(results_df))]) pve_snp pve_weight1 pve_weight2 pve_weight3
0.397402438 0.046985749 0.019935637 0.006963255 #store results for figure
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS G - Ind Var",
count=results_df$n_detected_comb_pip-results_df$n_detected_comb_pip_in_causal,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS G - Ind Var",
count=results_df$n_detected_comb_pip_in_causal,
ifcausal=T))Individual tissue analyses
For the cTWAS analysis, each tissue was analyzed individually and the results were combined.
results_dir <- "/project2/xinhe/shengqian/cTWAS/cTWAS_simulation/simulation_test_merge/"
runtag = "ukb-s80.45-liv_wb"
configtag <- 2
simutags <- paste(2, 1:5, sep = "-")
thin <- 0.1
sample_size <- 45000
PIP_threshold <- 0.8
results_df <- data.frame(simutag=as.character(),
n_causal_combined=as.integer(),
n_detected_weight1=as.integer(),
n_detected_in_causal_weight1=as.integer(),
n_detected_weight2=as.integer(),
n_detected_in_causal_weight2=as.integer(),
n_detected_weight3=as.integer(),
n_detected_in_causal_weight3=as.integer(),
n_detected_combined=as.integer(),
n_detected_in_causal_combined=as.integer())
for (i in 1:length(simutags)){
simutag <- simutags[i]
#load genes with true simulated effect
load(paste0(results_dir, runtag, "_simu", simutag, "-pheno.Rd"))
true_genes <- unlist(sapply(1:22, function(x){phenores$batch[[x]]$id.cgene}))
true_genes_combined <- unique(sapply(true_genes, function(x){unlist(strsplit(x, "[|]"))[1]}))
#number of causal genes
n_causal_combined <- length(true_genes_combined)
#load cTWAS results for weight1
ctwas_res <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR_weight1.susieIrss.txt"))
ctwas_gene_res <- ctwas_res[ctwas_res$type!="SNP",]
#number of genes with cTWAS PIP > threshold in weight1
ctwas_genes_weight1 <- ctwas_gene_res$id[ctwas_gene_res$susie_pip > PIP_threshold]
n_ctwas_genes_weight1 <- length(ctwas_genes_weight1)
n_causal_detected_weight1 <- sum(ctwas_genes_weight1 %in% true_genes_combined)
#load cTWAS results for weight2
ctwas_res <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR_weight2.susieIrss.txt"))
ctwas_gene_res <- ctwas_res[ctwas_res$type!="SNP",]
#number of genes with cTWAS PIP > threshold in weight1
ctwas_genes_weight2 <- ctwas_gene_res$id[ctwas_gene_res$susie_pip > PIP_threshold]
n_ctwas_genes_weight2 <- length(ctwas_genes_weight2)
n_causal_detected_weight2 <- sum(ctwas_genes_weight2 %in% true_genes_combined)
#load cTWAS results for weight3
ctwas_res <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR_weight3.susieIrss.txt"))
ctwas_gene_res <- ctwas_res[ctwas_res$type!="SNP",]
#number of genes with cTWAS PIP > threshold in weight3
ctwas_genes_weight3 <- ctwas_gene_res$id[ctwas_gene_res$susie_pip > PIP_threshold]
n_ctwas_genes_weight3 <- length(ctwas_genes_weight3)
n_causal_detected_weight3 <- sum(ctwas_genes_weight3 %in% true_genes_combined)
#combined analysis
ctwas_genes_combined <- unique(c(ctwas_genes_weight1, ctwas_genes_weight2, ctwas_genes_weight3))
n_ctwas_genes_combined <- length(ctwas_genes_combined)
n_causal_detected_combined <- sum(ctwas_genes_combined %in% true_genes_combined)
results_current <- data.frame(simutag=as.character(simutag),
n_causal_combined=as.integer(n_causal_combined),
n_detected_weight1=as.integer(n_ctwas_genes_weight1),
n_detected_in_causal_weight1=as.integer(n_causal_detected_weight1),
n_detected_weight2=as.integer(n_ctwas_genes_weight2),
n_detected_in_causal_weight2=as.integer(n_causal_detected_weight2),
n_detected_weight3=as.integer(n_ctwas_genes_weight3),
n_detected_in_causal_weight3=as.integer(n_causal_detected_weight3),
n_detected_combined=as.integer(n_ctwas_genes_combined),
n_detected_in_causal_combined=as.integer(n_causal_detected_combined))
results_df <- rbind(results_df, results_current)
}
#results using weight1
results_df[,c("simutag", colnames(results_df)[grep("weight1", colnames(results_df))])] simutag n_detected_weight1 n_detected_in_causal_weight1
1 2-1 55 42
2 2-2 74 57
3 2-3 50 32
4 2-4 49 35
5 2-5 54 41#mean percent causal using PIP > 0.8 for weight1
sum(results_df$n_detected_in_causal_weight1)/sum(results_df$n_detected_weight1)[1] 0.7340426#results using weight2
results_df[,c("simutag", colnames(results_df)[grep("weight2", colnames(results_df))])] simutag n_detected_weight2 n_detected_in_causal_weight2
1 2-1 25 12
2 2-2 52 31
3 2-3 35 23
4 2-4 42 28
5 2-5 33 17#mean percent causal using PIP > 0.8 for weight1
sum(results_df$n_detected_in_causal_weight2)/sum(results_df$n_detected_weight2)[1] 0.5935829#results using weight3
results_df[,c("simutag", colnames(results_df)[grep("weight3", colnames(results_df))])] simutag n_detected_weight3 n_detected_in_causal_weight3
1 2-1 23 11
2 2-2 27 13
3 2-3 18 5
4 2-4 19 10
5 2-5 30 12#mean percent causal using PIP > 0.8 for weight3
sum(results_df$n_detected_in_causal_weight3)/sum(results_df$n_detected_weight3)[1] 0.4358974#results using combined analysis
results_df[,c("simutag", colnames(results_df)[grep("combined", colnames(results_df))])] simutag n_causal_combined n_detected_combined n_detected_in_causal_combined
1 2-1 214 81 50
2 2-2 247 121 75
3 2-3 216 79 42
4 2-4 231 86 53
5 2-5 230 86 47#mean percent causal using PIP > 0.8 for weight3
sum(results_df$n_detected_in_causal_combined)/sum(results_df$n_detected_combined)[1] 0.589404#store results for figure
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight1",
count=results_df$n_detected_weight1-results_df$n_detected_in_causal_weight1,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight1",
count=results_df$n_detected_in_causal_weight1,
ifcausal=T))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight2",
count=results_df$n_detected_weight2-results_df$n_detected_in_causal_weight2,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight2",
count=results_df$n_detected_in_causal_weight2,
ifcausal=T))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight3",
count=results_df$n_detected_weight3-results_df$n_detected_in_causal_weight3,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight3",
count=results_df$n_detected_in_causal_weight3,
ifcausal=T))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Union",
count=results_df$n_detected_combined-results_df$n_detected_in_causal_combined,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Union",
count=results_df$n_detected_in_causal_combined,
ifcausal=T))Figure
For the cTWAS analysis, each tissue was analyzed individually and the results were combined.
plot_df$method <- factor(plot_df$method, levels=c("TWAS", "cTWAS G+T", "cTWAS G", "cTWAS G - Ind Var", "cTWAS Union", "cTWAS Weight1", "cTWAS Weight2", "cTWAS Weight3"))
library(ggpubr)
colset = c("#ebebeb", "#fb8072")
ggbarplot(plot_df,
x = "method",
y = "count",
add = "mean_se",
fill = "ifcausal",
legend = "none",
ylab="Count",
xlab="",
palette = colset) + grids(linetype = "dashed") + theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1))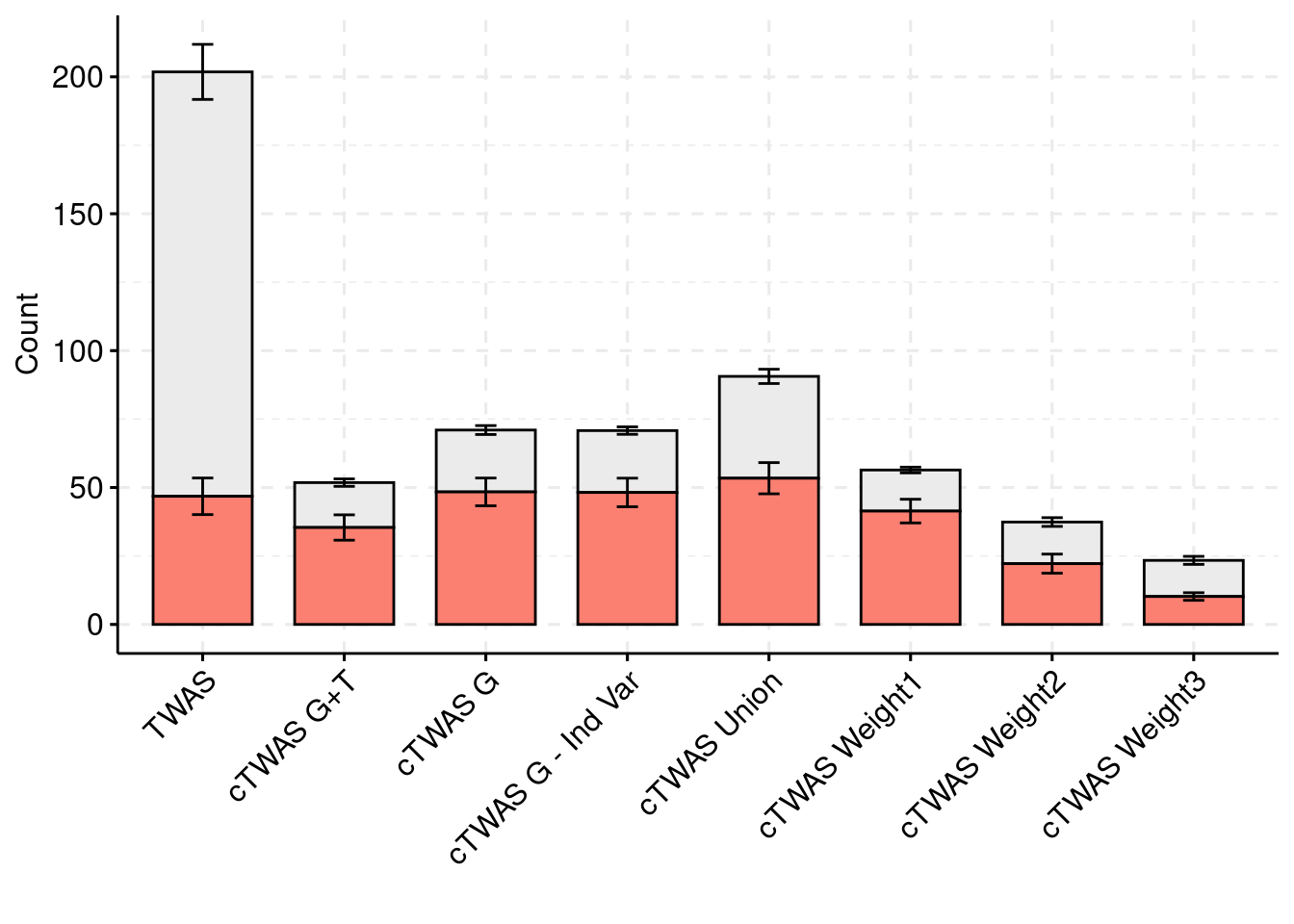
| Version | Author | Date |
|---|---|---|
| 9ed1bc7 | sq-96 | 2023-10-05 |
plot_df_2 <- plot_df
plot_df_2$method %in% c("TWAS", "cTWAS Union", "cTWAS Weight1", "cTWAS Weight2", "cTWAS Weight3") [1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[13] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE
[25] TRUE TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
[37] FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[49] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[61] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[73] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUEplot_df_2$ifcausal <- as.character(plot_df_2$ifcausal+1)
plot_df_2$count[plot_df_2$method=="cTWAS G" & plot_df_2$ifcausal=="2"] <- plot_df_2$count[plot_df_2$method=="cTWAS G" & plot_df_2$ifcausal=="2"] - plot_df_2$count[plot_df_2$method=="cTWAS G+T" & plot_df_2$ifcausal=="2"]
plot_df_2 <- rbind(plot_df_2, data.frame(simutag=simutags,
method="cTWAS G",
count=plot_df_2$count[plot_df_2$method=="cTWAS G+T" & plot_df_2$ifcausal=="2"],
ifcausal="3"))
plot_df_3 <- plot_df_2
plot_df_2 <- plot_df_2[plot_df_2$method %in% c("TWAS", "cTWAS Union", "cTWAS Weight1", "cTWAS Weight2", "cTWAS Weight3"),]
plot_df_2$method <- droplevels(plot_df_2$method)
levels(plot_df_2$method) <- c("TWAS", "cTWAS Union", "cTWAS Primary", "cTWAS Secondary", "cTWAS Null")
plot_df_2$ifcausal[plot_df_2$ifcausal=="1"] <- "FP"
plot_df_2$ifcausal[plot_df_2$ifcausal=="2"] <- "TP"
colset = c("#ebebeb", "#fb8072", "#8dd3c7")
ggbarplot(plot_df_2,
x = "method",
y = "count",
add = "mean_se",
fill = "ifcausal",
legend = "right",
ylab="Count",
xlab="",
palette = colset) + grids(linetype = "dashed") + theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1)) + guides(fill=guide_legend(title=""))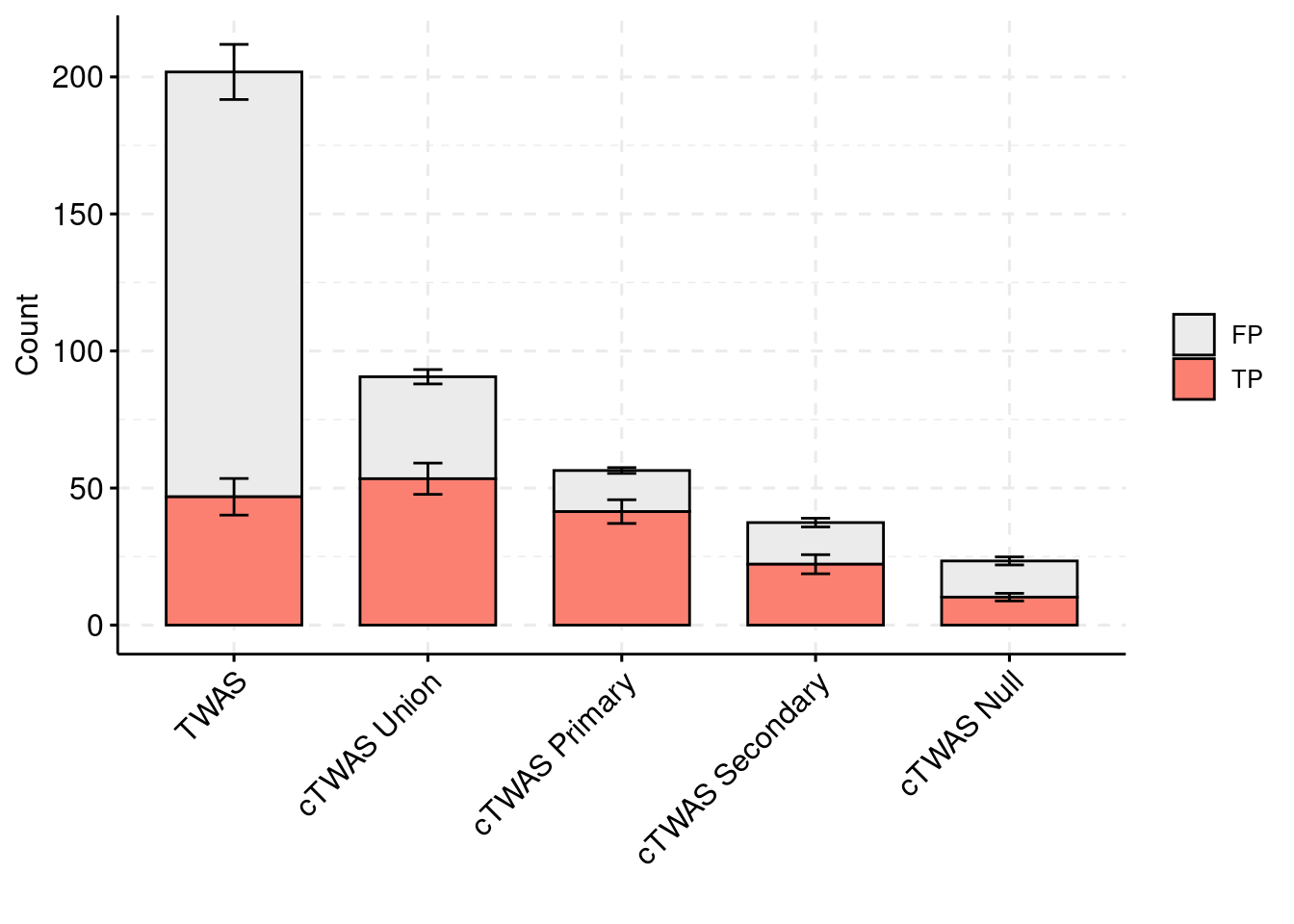
| Version | Author | Date |
|---|---|---|
| 9ed1bc7 | sq-96 | 2023-10-05 |
plot_df_3 <- plot_df_3[plot_df_3$method %in% c("cTWAS G", "cTWAS Union"),]
plot_df_3$method <- droplevels(plot_df_3$method)
levels(plot_df_3$method) <- c("cTWAS Joint", "cTWAS Union")
plot_df_3$method <- relevel(plot_df_3$method, "cTWAS Union")
plot_df_3$ifcausal[plot_df_3$ifcausal=="1"] <- "FP"
plot_df_3$ifcausal[plot_df_3$ifcausal=="2"] <- "TP"
plot_df_3$ifcausal[plot_df_3$ifcausal=="3"] <- "TP and Tissue Identified\nby Joint Analysis"
ggbarplot(plot_df_3,
x = "method",
y = "count",
add = "mean_se",
fill = "ifcausal",
legend = "right",
ylab="Count",
xlab="",
palette = colset) + grids(linetype = "dashed") + theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1)) + guides(fill=guide_legend(title=""))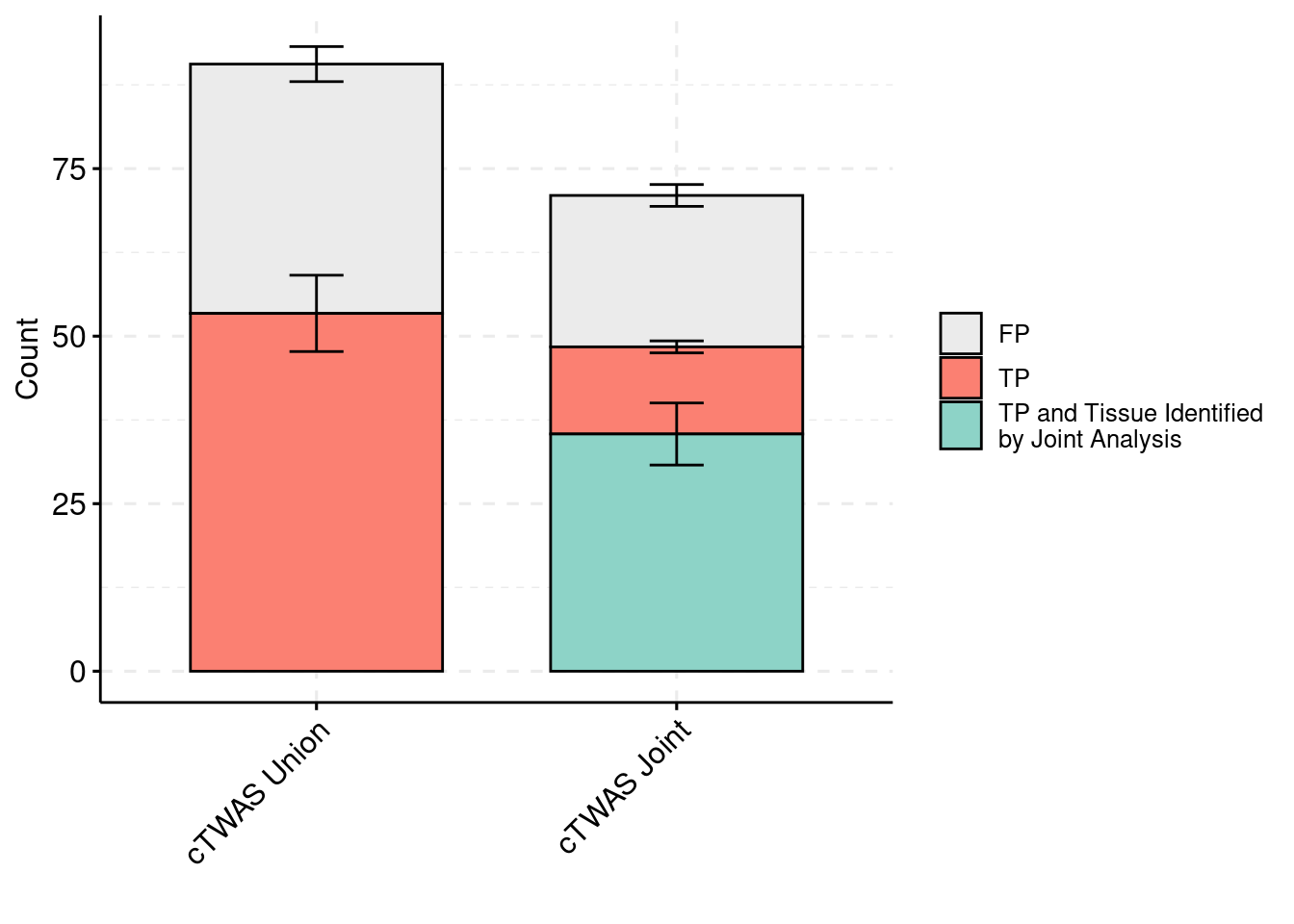
| Version | Author | Date |
|---|---|---|
| 9ed1bc7 | sq-96 | 2023-10-05 |
Simulation 3: One causal tissue
Separate effect size parameters
For the cTWAS analysis, each tissue had its own prior inclusion parameter and effect size parameter.
results_dir <- "/project2/xinhe/shengqian/cTWAS/cTWAS_simulation/simulation_test_merge/"
runtag = "ukb-s80.45-liv_wb"
configtag <- 2
simutags <- paste(3, 1:5, sep = "-")
thin <- 0.1
sample_size <- 45000
PIP_threshold <- 0.8
results_df <- data.frame(simutag=as.character(),
n_causal=as.integer(),
n_causal_combined=as.integer(),
n_detected_pip=as.integer(),
n_detected_pip_in_causal=as.integer(),
n_detected_comb_pip=as.integer(),
n_detected_comb_pip_in_causal=as.integer(),
pve_snp=as.numeric(),
pve_weight1=as.numeric(),
pve_weight2=as.numeric(),
pve_weight3=as.numeric(),
prior_weight1=as.numeric(),
prior_weight2=as.numeric(),
prior_weight3=as.numeric(),
prior_var_snp=as.numeric(),
prior_var_weight1=as.numeric(),
prior_var_weight2=as.numeric(),
prior_var_weight3=as.numeric())
for (i in 1:length(simutags)){
simutag <- simutags[i]
#load genes with true simulated effect
load(paste0(results_dir, runtag, "_simu", simutag, "-pheno.Rd"))
true_genes <- unlist(sapply(1:22, function(x){phenores$batch[[x]]$id.cgene}))
true_genes_combined <- unique(sapply(true_genes, function(x){unlist(strsplit(x, "[|]"))[1]}))
#load cTWAS results
ctwas_res <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR.susieIrss.txt"))
ctwas_gene_res <- ctwas_res[ctwas_res$type!="SNP",]
#number of causal genes
n_causal <- length(true_genes)
n_causal_combined <- length(true_genes_combined)
#number of gene+tissue combinations with cTWAS PIP > threshold
n_ctwas_genes <- sum(ctwas_gene_res$susie_pip > PIP_threshold)
#number of cTWAS genes that are causal
n_causal_detected <- sum(ctwas_gene_res$id[ctwas_gene_res$susie_pip > PIP_threshold] %in% true_genes)
#collapse gene+tissues to genes and compute combined PIP
ctwas_gene_res$gene <- sapply(ctwas_gene_res$id, function(x){unlist(strsplit(x,"[|]"))[1]})
ctwas_gene_res_combined <- aggregate(ctwas_gene_res$susie_pip, by=list(ctwas_gene_res$gene), FUN=sum)
colnames(ctwas_gene_res_combined) <- c("gene", "pip_combined")
#number of genes with combined PIP > threshold
n_ctwas_genes_combined <- sum(ctwas_gene_res_combined$pip_combined > PIP_threshold)
#number of cTWAS genes using combined PIP that are causal
n_causal_detected_combined <- sum(ctwas_gene_res_combined$gene[ctwas_gene_res_combined$pip_combined > PIP_threshold] %in% true_genes_combined)
#collect number of SNPs analyzed by cTWAS
ctwas_res_s1 <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR.s1.susieIrss.txt"))
n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
rm(ctwas_res_s1)
#load estimated parameters
load(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR.s2.susieIrssres.Rd"))
#estimated group prior (all iterations)
estimated_group_prior_all <- group_prior_rec
estimated_group_prior_all["SNP",] <- estimated_group_prior_all["SNP",]*thin #adjust parameter to account for thin argument
#estimated group prior variance (all iterations)
estimated_group_prior_var_all <- group_prior_var_rec
#set group size
group_size <- c(table(ctwas_gene_res$type), structure(n_snps, names="SNP"))
group_size <- group_size[rownames(estimated_group_prior_all)]
#estimated group PVE (all iterations)
estimated_group_pve_all <- estimated_group_prior_var_all*estimated_group_prior_all*group_size/sample_size #check PVE calculation
results_current <- data.frame(simutag=as.character(simutag),
n_causal=as.integer(n_causal),
n_causal_combined=as.integer(n_causal_combined),
n_detected_pip=as.integer(n_ctwas_genes),
n_detected_pip_in_causal=as.integer(n_causal_detected),
n_detected_comb_pip=as.integer(n_ctwas_genes_combined),
n_detected_comb_pip_in_causal=as.integer(n_causal_detected_combined),
pve_snp=as.numeric(rev(estimated_group_pve_all["SNP",])[1]),
pve_weight1=as.numeric(rev(estimated_group_pve_all["Liver",])[1]),
pve_weight2=as.numeric(rev(estimated_group_pve_all["Whole_Blood",])[1]),
pve_weight3=as.numeric(rev(estimated_group_pve_all["Brain_Cerebellum",])[1]),
prior_snp=as.numeric(rev(estimated_group_prior_var_all["SNP",])[1]),
prior_weight1=as.numeric(rev(estimated_group_prior_all["Liver",])[1]),
prior_weight2=as.numeric(rev(estimated_group_prior_all["Whole_Blood",])[1]),
prior_weight3=as.numeric(rev(estimated_group_prior_all["Brain_Cerebellum",])[1]),
prior_var_snp=as.numeric(rev(estimated_group_prior_var_all["SNP",])[1]),
prior_var_weight1=as.numeric(rev(estimated_group_prior_var_all["Liver",])[1]),
prior_var_weight2=as.numeric(rev(estimated_group_prior_var_all["Whole_Blood",])[1]),
prior_var_weight3=as.numeric(rev(estimated_group_prior_var_all["Brain_Cerebellum",])[1]))
results_df <- rbind(results_df, results_current)
}
#results using PIP threshold (gene+tissue)
results_df[,c("simutag", "n_causal", "n_detected_pip", "n_detected_pip_in_causal")] simutag n_causal n_detected_pip n_detected_pip_in_causal
1 3-1 118 40 30
2 3-2 95 51 34
3 3-3 116 36 26
4 3-4 117 47 37
5 3-5 113 48 37#mean percent causal using PIP > 0.8
sum(results_df$n_detected_pip_in_causal)/sum(results_df$n_detected_pip)[1] 0.7387387#results using combined PIP threshold
results_df[,c("simutag", "n_causal_combined", "n_detected_comb_pip", "n_detected_comb_pip_in_causal")] simutag n_causal_combined n_detected_comb_pip n_detected_comb_pip_in_causal
1 3-1 118 46 33
2 3-2 95 51 34
3 3-3 116 54 34
4 3-4 117 60 41
5 3-5 113 55 41#mean percent causal using combined PIP > 0.8
sum(results_df$n_detected_comb_pip_in_causal)/sum(results_df$n_detected_comb_pip)[1] 0.6879699#prior inclusion and mean prior inclusion
results_df[,c(which(colnames(results_df)=="simutag"), setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df))))] simutag prior_snp prior_weight1 prior_weight2 prior_weight3
1 3-1 15.26854 0.009960057 0.0029193316 0.0006996409
2 3-2 14.21774 0.015921227 0.0005239939 0.0001075432
3 3-3 13.00977 0.011268497 0.0045913157 0.0039885322
4 3-4 13.40018 0.020094595 0.0024137731 0.0048414159
5 3-5 13.04042 0.017810800 0.0036165052 0.0014596020colMeans(results_df[,setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df)))]) prior_snp prior_weight1 prior_weight2 prior_weight3
13.787332323 0.015011035 0.002812984 0.002219347 #prior variance and mean prior variance
results_df[,c(which(colnames(results_df)=="simutag"), grep("prior_var", names(results_df)))] simutag prior_var_snp prior_var_weight1 prior_var_weight2 prior_var_weight3
1 3-1 15.26854 31.33725 8.544893 1.151033
2 3-2 14.21774 21.99653 2.170788 1.777382
3 3-3 13.00977 29.18026 12.599922 8.153568
4 3-4 13.40018 16.87536 10.573233 6.239371
5 3-5 13.04042 23.99469 2.553775 15.738786colMeans(results_df[,grep("prior_var", names(results_df))]) prior_var_snp prior_var_weight1 prior_var_weight2 prior_var_weight3
13.787332 24.676817 7.288522 6.612028 #PVE and mean PVE
results_df[,c(which(colnames(results_df)=="simutag"), grep("pve", names(results_df)))] simutag pve_snp pve_weight1 pve_weight2 pve_weight3
1 3-1 0.3902628 0.05129185 0.0045289716 1.574470e-04
2 3-2 0.4134510 0.05755146 0.0002065158 3.737101e-05
3 3-3 0.4120362 0.05403570 0.0105030290 6.358171e-03
4 3-4 0.4290854 0.05572602 0.0046335493 5.905881e-03
5 3-5 0.3937832 0.07023025 0.0016768003 4.491352e-03colMeans(results_df[,grep("pve", names(results_df))]) pve_snp pve_weight1 pve_weight2 pve_weight3
0.407723705 0.057767056 0.004309773 0.003390045 #store results for figure
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS G - Ind Var",
count=results_df$n_detected_comb_pip-results_df$n_detected_comb_pip_in_causal,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS G - Ind Var",
count=results_df$n_detected_comb_pip_in_causal,
ifcausal=T))Individual tissue analyses
For the cTWAS analysis, each tissue was analyzed individually and the results were combined.
results_dir <- "/project2/xinhe/shengqian/cTWAS/cTWAS_simulation/simulation_test_merge/"
runtag = "ukb-s80.45-liv_wb"
configtag <- 2
simutags <- paste(3, 1:5, sep = "-")
thin <- 0.1
sample_size <- 45000
PIP_threshold <- 0.8
results_df <- data.frame(simutag=as.character(),
n_causal_combined=as.integer(),
n_detected_weight1=as.integer(),
n_detected_in_causal_weight1=as.integer(),
n_detected_weight2=as.integer(),
n_detected_in_causal_weight2=as.integer(),
n_detected_weight3=as.integer(),
n_detected_in_causal_weight3=as.integer(),
n_detected_combined=as.integer(),
n_detected_in_causal_combined=as.integer())
for (i in 1:length(simutags)){
simutag <- simutags[i]
#load genes with true simulated effect
load(paste0(results_dir, runtag, "_simu", simutag, "-pheno.Rd"))
true_genes <- unlist(sapply(1:22, function(x){phenores$batch[[x]]$id.cgene}))
true_genes_combined <- unique(sapply(true_genes, function(x){unlist(strsplit(x, "[|]"))[1]}))
#number of causal genes
n_causal_combined <- length(true_genes_combined)
#load cTWAS results for weight1
ctwas_res <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR_weight1.susieIrss.txt"))
ctwas_gene_res <- ctwas_res[ctwas_res$type!="SNP",]
#number of genes with cTWAS PIP > threshold in weight1
ctwas_genes_weight1 <- ctwas_gene_res$id[ctwas_gene_res$susie_pip > PIP_threshold]
n_ctwas_genes_weight1 <- length(ctwas_genes_weight1)
n_causal_detected_weight1 <- sum(ctwas_genes_weight1 %in% true_genes_combined)
#load cTWAS results for weight2
ctwas_res <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR_weight2.susieIrss.txt"))
ctwas_gene_res <- ctwas_res[ctwas_res$type!="SNP",]
#number of genes with cTWAS PIP > threshold in weight1
ctwas_genes_weight2 <- ctwas_gene_res$id[ctwas_gene_res$susie_pip > PIP_threshold]
n_ctwas_genes_weight2 <- length(ctwas_genes_weight2)
n_causal_detected_weight2 <- sum(ctwas_genes_weight2 %in% true_genes_combined)
#load cTWAS results for weight3
ctwas_res <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR_weight3.susieIrss.txt"))
ctwas_gene_res <- ctwas_res[ctwas_res$type!="SNP",]
#number of genes with cTWAS PIP > threshold in weight3
ctwas_genes_weight3 <- ctwas_gene_res$id[ctwas_gene_res$susie_pip > PIP_threshold]
n_ctwas_genes_weight3 <- length(ctwas_genes_weight3)
n_causal_detected_weight3 <- sum(ctwas_genes_weight3 %in% true_genes_combined)
#combined analysis
ctwas_genes_combined <- unique(c(ctwas_genes_weight1, ctwas_genes_weight2, ctwas_genes_weight3))
n_ctwas_genes_combined <- length(ctwas_genes_combined)
n_causal_detected_combined <- sum(ctwas_genes_combined %in% true_genes_combined)
results_current <- data.frame(simutag=as.character(simutag),
n_causal_combined=as.integer(n_causal_combined),
n_detected_weight1=as.integer(n_ctwas_genes_weight1),
n_detected_in_causal_weight1=as.integer(n_causal_detected_weight1),
n_detected_weight2=as.integer(n_ctwas_genes_weight2),
n_detected_in_causal_weight2=as.integer(n_causal_detected_weight2),
n_detected_weight3=as.integer(n_ctwas_genes_weight3),
n_detected_in_causal_weight3=as.integer(n_causal_detected_weight3),
n_detected_combined=as.integer(n_ctwas_genes_combined),
n_detected_in_causal_combined=as.integer(n_causal_detected_combined))
results_df <- rbind(results_df, results_current)
}
#results using weight1
results_df[,c("simutag", colnames(results_df)[grep("weight1", colnames(results_df))])] simutag n_detected_weight1 n_detected_in_causal_weight1
1 3-1 43 33
2 3-2 51 34
3 3-3 54 35
4 3-4 56 42
5 3-5 51 40#mean percent causal using PIP > 0.8 for weight1
sum(results_df$n_detected_in_causal_weight1)/sum(results_df$n_detected_weight1)[1] 0.7215686#results using weight2
results_df[,c("simutag", colnames(results_df)[grep("weight2", colnames(results_df))])] simutag n_detected_weight2 n_detected_in_causal_weight2
1 3-1 11 4
2 3-2 12 5
3 3-3 19 9
4 3-4 16 9
5 3-5 15 8#mean percent causal using PIP > 0.8 for weight1
sum(results_df$n_detected_in_causal_weight2)/sum(results_df$n_detected_weight2)[1] 0.4794521#results using weight3
results_df[,c("simutag", colnames(results_df)[grep("weight3", colnames(results_df))])] simutag n_detected_weight3 n_detected_in_causal_weight3
1 3-1 3 2
2 3-2 7 5
3 3-3 14 3
4 3-4 21 10
5 3-5 8 5#mean percent causal using PIP > 0.8 for weight3
sum(results_df$n_detected_in_causal_weight3)/sum(results_df$n_detected_weight3)[1] 0.4716981#results using combined analysis
results_df[,c("simutag", colnames(results_df)[grep("combined", colnames(results_df))])] simutag n_causal_combined n_detected_combined n_detected_in_causal_combined
1 3-1 118 49 33
2 3-2 95 58 34
3 3-3 116 71 35
4 3-4 117 73 46
5 3-5 113 58 40#mean percent causal using PIP > 0.8 for weight3
sum(results_df$n_detected_in_causal_combined)/sum(results_df$n_detected_combined)[1] 0.6084142#store results for figure
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight1",
count=results_df$n_detected_weight1-results_df$n_detected_in_causal_weight1,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight1",
count=results_df$n_detected_in_causal_weight1,
ifcausal=T))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight2",
count=results_df$n_detected_weight2-results_df$n_detected_in_causal_weight2,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight2",
count=results_df$n_detected_in_causal_weight2,
ifcausal=T))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight3",
count=results_df$n_detected_weight3-results_df$n_detected_in_causal_weight3,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight3",
count=results_df$n_detected_in_causal_weight3,
ifcausal=T))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Union",
count=results_df$n_detected_combined-results_df$n_detected_in_causal_combined,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Union",
count=results_df$n_detected_in_causal_combined,
ifcausal=T))Figure
For the cTWAS analysis, each tissue was analyzed individually and the results were combined.
plot_df$method <- factor(plot_df$method, levels=c("TWAS", "cTWAS G+T", "cTWAS G", "cTWAS G - Ind Var", "cTWAS Union", "cTWAS Weight1", "cTWAS Weight2", "cTWAS Weight3"))
library(ggpubr)
colset = c("#ebebeb", "#fb8072")
ggbarplot(plot_df,
x = "method",
y = "count",
add = "mean_se",
fill = "ifcausal",
legend = "none",
ylab="Count",
xlab="",
palette = colset) + grids(linetype = "dashed") + theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1))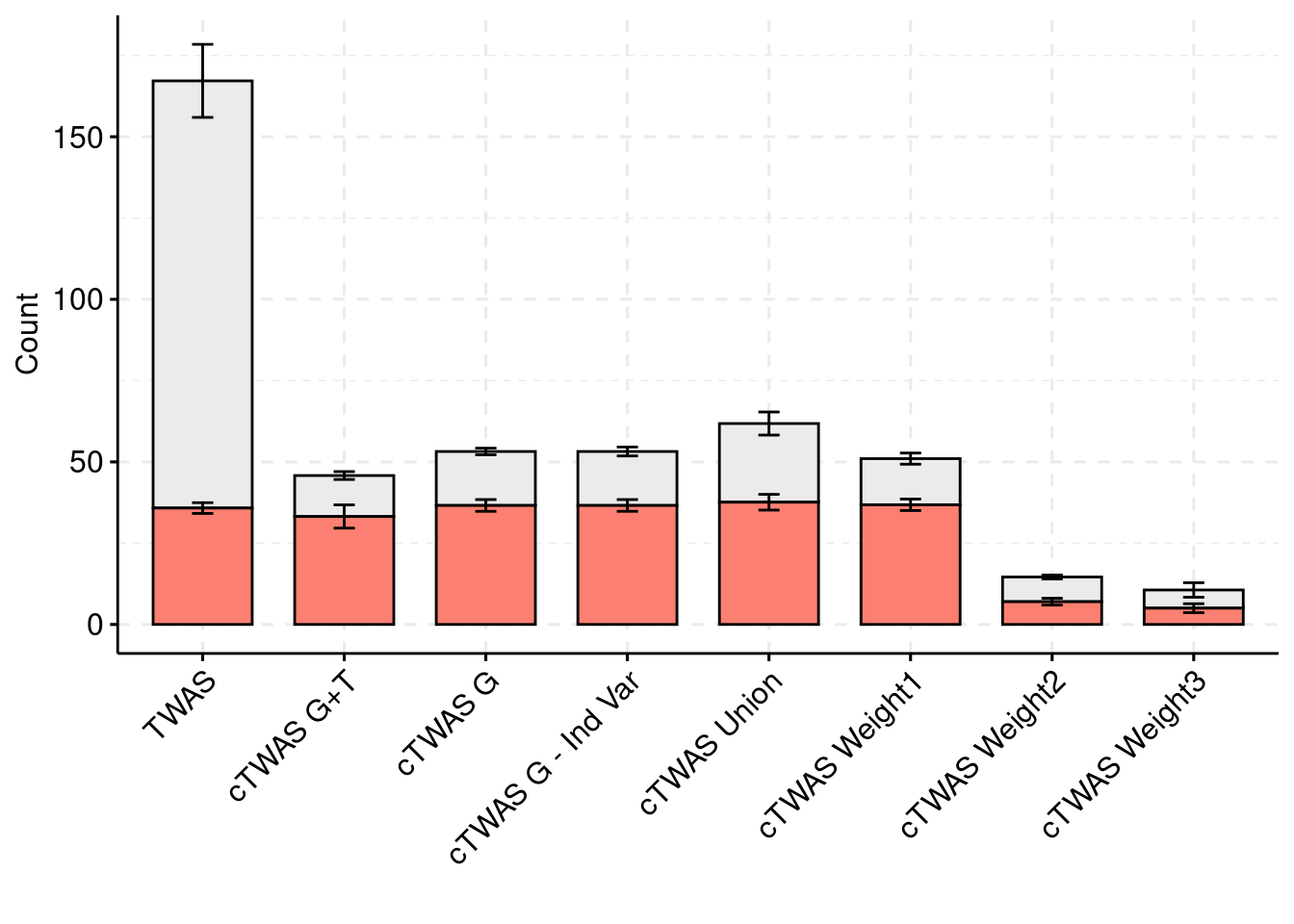
| Version | Author | Date |
|---|---|---|
| 9ed1bc7 | sq-96 | 2023-10-05 |
plot_df_2 <- plot_df
plot_df_2$method %in% c("TWAS", "cTWAS Union", "cTWAS Weight1", "cTWAS Weight2", "cTWAS Weight3") [1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[13] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE
[25] TRUE TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
[37] FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[49] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[61] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[73] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUEplot_df_2$ifcausal <- as.character(plot_df_2$ifcausal+1)
plot_df_2$count[plot_df_2$method=="cTWAS G" & plot_df_2$ifcausal=="2"] <- plot_df_2$count[plot_df_2$method=="cTWAS G" & plot_df_2$ifcausal=="2"] - plot_df_2$count[plot_df_2$method=="cTWAS G+T" & plot_df_2$ifcausal=="2"]
plot_df_2 <- rbind(plot_df_2, data.frame(simutag=simutags,
method="cTWAS G",
count=plot_df_2$count[plot_df_2$method=="cTWAS G+T" & plot_df_2$ifcausal=="2"],
ifcausal="3"))
plot_df_3 <- plot_df_2
plot_df_2 <- plot_df_2[plot_df_2$method %in% c("TWAS", "cTWAS Union", "cTWAS Weight1", "cTWAS Weight2", "cTWAS Weight3"),]
plot_df_2$method <- droplevels(plot_df_2$method)
levels(plot_df_2$method) <- c("TWAS", "cTWAS Union", "cTWAS Primary", "cTWAS Secondary", "cTWAS Null")
plot_df_2$ifcausal[plot_df_2$ifcausal=="1"] <- "FP"
plot_df_2$ifcausal[plot_df_2$ifcausal=="2"] <- "TP"
colset = c("#ebebeb", "#fb8072", "#8dd3c7")
ggbarplot(plot_df_2,
x = "method",
y = "count",
add = "mean_se",
fill = "ifcausal",
legend = "right",
ylab="Count",
xlab="",
palette = colset) + grids(linetype = "dashed") + theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1)) + guides(fill=guide_legend(title=""))
| Version | Author | Date |
|---|---|---|
| 9ed1bc7 | sq-96 | 2023-10-05 |
plot_df_3 <- plot_df_3[plot_df_3$method %in% c("cTWAS G", "cTWAS Union"),]
plot_df_3$method <- droplevels(plot_df_3$method)
levels(plot_df_3$method) <- c("cTWAS Joint", "cTWAS Union")
plot_df_3$method <- relevel(plot_df_3$method, "cTWAS Union")
plot_df_3$ifcausal[plot_df_3$ifcausal=="1"] <- "FP"
plot_df_3$ifcausal[plot_df_3$ifcausal=="2"] <- "TP"
plot_df_3$ifcausal[plot_df_3$ifcausal=="3"] <- "TP and Tissue Identified\nby Joint Analysis"
ggbarplot(plot_df_3,
x = "method",
y = "count",
add = "mean_se",
fill = "ifcausal",
legend = "right",
ylab="Count",
xlab="",
palette = colset) + grids(linetype = "dashed") + theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1)) + guides(fill=guide_legend(title=""))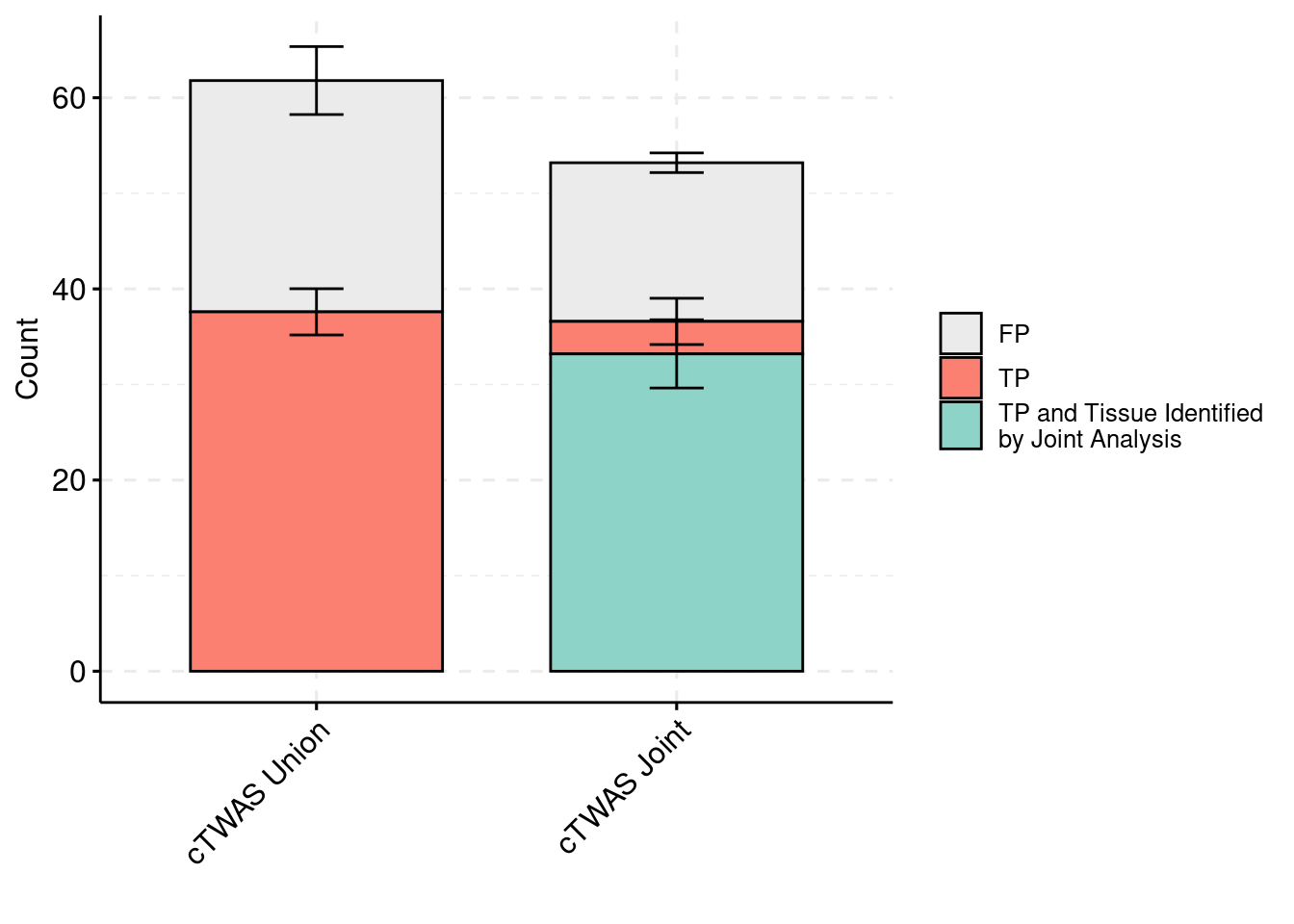
| Version | Author | Date |
|---|---|---|
| 9ed1bc7 | sq-96 | 2023-10-05 |
Simulation 4: No causal tissues
Separate effect size parameters
For the cTWAS analysis, each tissue had its own prior inclusion parameter end effect size parameter.
results_dir <- "/project2/xinhe/shengqian/cTWAS/cTWAS_simulation/simulation_test_merge/"
runtag = "ukb-s80.45-liv_wb"
configtag <- 2
simutags <- paste(4, 1:5, sep = "-")
thin <- 0.1
sample_size <- 45000
PIP_threshold <- 0.8
results_df <- data.frame(simutag=as.character(),
n_causal=as.integer(),
n_causal_combined=as.integer(),
n_detected_pip=as.integer(),
n_detected_pip_in_causal=as.integer(),
n_detected_comb_pip=as.integer(),
n_detected_comb_pip_in_causal=as.integer(),
pve_snp=as.numeric(),
pve_weight1=as.numeric(),
pve_weight2=as.numeric(),
pve_weight3=as.numeric(),
prior_weight1=as.numeric(),
prior_weight2=as.numeric(),
prior_weight3=as.numeric(),
prior_var_snp=as.numeric(),
prior_var_weight1=as.numeric(),
prior_var_weight2=as.numeric(),
prior_var_weight3=as.numeric())
for (i in 1:length(simutags)){
simutag <- simutags[i]
#load genes with true simulated effect
load(paste0(results_dir, runtag, "_simu", simutag, "-pheno.Rd"))
true_genes <- unlist(sapply(1:22, function(x){phenores$batch[[x]]$id.cgene}))
true_genes_combined <- unique(sapply(true_genes, function(x){unlist(strsplit(x, "[|]"))[1]}))
#load cTWAS results
ctwas_res <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR.susieIrss.txt"))
ctwas_gene_res <- ctwas_res[ctwas_res$type!="SNP",]
#number of causal genes
n_causal <- length(true_genes)
n_causal_combined <- length(true_genes_combined)
#number of gene+tissue combinations with cTWAS PIP > threshold
n_ctwas_genes <- sum(ctwas_gene_res$susie_pip > PIP_threshold)
#number of cTWAS genes that are causal
n_causal_detected <- sum(ctwas_gene_res$id[ctwas_gene_res$susie_pip > PIP_threshold] %in% true_genes)
#collapse gene+tissues to genes and compute combined PIP
ctwas_gene_res$gene <- sapply(ctwas_gene_res$id, function(x){unlist(strsplit(x,"[|]"))[1]})
ctwas_gene_res_combined <- aggregate(ctwas_gene_res$susie_pip, by=list(ctwas_gene_res$gene), FUN=sum)
colnames(ctwas_gene_res_combined) <- c("gene", "pip_combined")
#number of genes with combined PIP > threshold
n_ctwas_genes_combined <- sum(ctwas_gene_res_combined$pip_combined > PIP_threshold)
#number of cTWAS genes using combined PIP that are causal
n_causal_detected_combined <- sum(ctwas_gene_res_combined$gene[ctwas_gene_res_combined$pip_combined > PIP_threshold] %in% true_genes_combined)
#collect number of SNPs analyzed by cTWAS
ctwas_res_s1 <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR.s1.susieIrss.txt"))
n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
rm(ctwas_res_s1)
#load estimated parameters
load(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR.s2.susieIrssres.Rd"))
#estimated group prior (all iterations)
estimated_group_prior_all <- group_prior_rec
estimated_group_prior_all["SNP",] <- estimated_group_prior_all["SNP",]*thin #adjust parameter to account for thin argument
#estimated group prior variance (all iterations)
estimated_group_prior_var_all <- group_prior_var_rec
#set group size
group_size <- c(table(ctwas_gene_res$type), structure(n_snps, names="SNP"))
group_size <- group_size[rownames(estimated_group_prior_all)]
#estimated group PVE (all iterations)
estimated_group_pve_all <- estimated_group_prior_var_all*estimated_group_prior_all*group_size/sample_size #check PVE calculation
results_current <- data.frame(simutag=as.character(simutag),
n_causal=as.integer(n_causal),
n_causal_combined=as.integer(n_causal_combined),
n_detected_pip=as.integer(n_ctwas_genes),
n_detected_pip_in_causal=as.integer(n_causal_detected),
n_detected_comb_pip=as.integer(n_ctwas_genes_combined),
n_detected_comb_pip_in_causal=as.integer(n_causal_detected_combined),
pve_snp=as.numeric(rev(estimated_group_pve_all["SNP",])[1]),
pve_weight1=as.numeric(rev(estimated_group_pve_all["Liver",])[1]),
pve_weight2=as.numeric(rev(estimated_group_pve_all["Whole_Blood",])[1]),
pve_weight3=as.numeric(rev(estimated_group_pve_all["Brain_Cerebellum",])[1]),
prior_snp=as.numeric(rev(estimated_group_prior_var_all["SNP",])[1]),
prior_weight1=as.numeric(rev(estimated_group_prior_all["Liver",])[1]),
prior_weight2=as.numeric(rev(estimated_group_prior_all["Whole_Blood",])[1]),
prior_weight3=as.numeric(rev(estimated_group_prior_all["Brain_Cerebellum",])[1]),
prior_var_snp=as.numeric(rev(estimated_group_prior_var_all["SNP",])[1]),
prior_var_weight1=as.numeric(rev(estimated_group_prior_var_all["Liver",])[1]),
prior_var_weight2=as.numeric(rev(estimated_group_prior_var_all["Whole_Blood",])[1]),
prior_var_weight3=as.numeric(rev(estimated_group_prior_var_all["Brain_Cerebellum",])[1]))
results_df <- rbind(results_df, results_current)
}
#results using PIP threshold (gene+tissue)
results_df[,c("simutag", "n_causal", "n_detected_pip", "n_detected_pip_in_causal")] simutag n_causal n_detected_pip n_detected_pip_in_causal
1 4-1 0 6 0
2 4-2 0 0 0
3 4-3 0 0 0
4 4-4 0 3 0
5 4-5 0 5 0#mean percent causal using PIP > 0.8
sum(results_df$n_detected_pip_in_causal)/sum(results_df$n_detected_pip)[1] 0#results using combined PIP threshold
results_df[,c("simutag", "n_causal_combined", "n_detected_comb_pip", "n_detected_comb_pip_in_causal")] simutag n_causal_combined n_detected_comb_pip n_detected_comb_pip_in_causal
1 4-1 0 6 0
2 4-2 0 0 0
3 4-3 0 3 0
4 4-4 0 4 0
5 4-5 0 5 0#mean percent causal using combined PIP > 0.8
sum(results_df$n_detected_comb_pip_in_causal)/sum(results_df$n_detected_comb_pip)[1] 0#prior inclusion and mean prior inclusion
results_df[,c(which(colnames(results_df)=="simutag"), setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df))))] simutag prior_snp prior_weight1 prior_weight2 prior_weight3
1 4-1 12.58359 0.0017288204 0.0030204233 0.003619867
2 4-2 13.08410 0.0005832831 0.0014533136 0.001222175
3 4-3 12.06660 0.0037554283 0.0009381176 0.002609262
4 4-4 11.14881 0.0073686855 0.0013821132 0.000845720
5 4-5 12.07505 0.0047356209 0.0026851585 0.001770055colMeans(results_df[,setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df)))]) prior_snp prior_weight1 prior_weight2 prior_weight3
12.191632680 0.003634368 0.001895825 0.002013416 #prior variance and mean prior variance
results_df[,c(which(colnames(results_df)=="simutag"), grep("prior_var", names(results_df)))] simutag prior_var_snp prior_var_weight1 prior_var_weight2 prior_var_weight3
1 4-1 12.58359 0.6479737 28.946442 0.4261434
2 4-2 13.08410 1.3718846 2.020467 38.9870059
3 4-3 12.06660 4.0111067 32.042210 3.7671720
4 4-4 11.14881 3.1822072 37.942118 0.8168949
5 4-5 12.07505 1.3471771 20.428115 0.6388463colMeans(results_df[,grep("prior_var", names(results_df))]) prior_var_snp prior_var_weight1 prior_var_weight2 prior_var_weight3
12.191633 2.112070 24.275870 8.927212 #PVE and mean PVE
results_df[,c(which(colnames(results_df)=="simutag"), grep("pve", names(results_df)))] simutag pve_snp pve_weight1 pve_weight2 pve_weight3
1 4-1 0.3776241 0.0001840911 0.0158734940 0.0003015920
2 4-2 0.4258038 0.0001314991 0.0005331148 0.0093159007
3 4-3 0.4219170 0.0024754226 0.0054574440 0.0019217837
4 4-4 0.3769374 0.0038534004 0.0095208286 0.0001350717
5 4-5 0.4087371 0.0010484007 0.0099588172 0.0002210826colMeans(results_df[,grep("pve", names(results_df))]) pve_snp pve_weight1 pve_weight2 pve_weight3
0.402203910 0.001538563 0.008268740 0.002379086 #store results for figure
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS G - Ind Var",
count=results_df$n_detected_comb_pip-results_df$n_detected_comb_pip_in_causal,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS G - Ind Var",
count=results_df$n_detected_comb_pip_in_causal,
ifcausal=T))Individual tissue analyses
For the cTWAS analysis, each tissue was analyzed individually and the results were combined.
results_dir <- "/project2/xinhe/shengqian/cTWAS/cTWAS_simulation/simulation_test_merge/"
runtag = "ukb-s80.45-liv_wb"
configtag <- 2
simutags <- paste(4, 1:5, sep = "-")
thin <- 0.1
sample_size <- 45000
PIP_threshold <- 0.8
results_df <- data.frame(simutag=as.character(),
n_causal_combined=as.integer(),
n_detected_weight1=as.integer(),
n_detected_in_causal_weight1=as.integer(),
n_detected_weight2=as.integer(),
n_detected_in_causal_weight2=as.integer(),
n_detected_weight3=as.integer(),
n_detected_in_causal_weight3=as.integer(),
n_detected_combined=as.integer(),
n_detected_in_causal_combined=as.integer())
for (i in 1:length(simutags)){
simutag <- simutags[i]
#load genes with true simulated effect
load(paste0(results_dir, runtag, "_simu", simutag, "-pheno.Rd"))
true_genes <- unlist(sapply(1:22, function(x){phenores$batch[[x]]$id.cgene}))
true_genes_combined <- unique(sapply(true_genes, function(x){unlist(strsplit(x, "[|]"))[1]}))
#number of causal genes
n_causal_combined <- length(true_genes_combined)
#load cTWAS results for weight1
ctwas_res <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR_weight1.susieIrss.txt"))
ctwas_gene_res <- ctwas_res[ctwas_res$type!="SNP",]
#number of genes with cTWAS PIP > threshold in weight1
ctwas_genes_weight1 <- ctwas_gene_res$id[ctwas_gene_res$susie_pip > PIP_threshold]
n_ctwas_genes_weight1 <- length(ctwas_genes_weight1)
n_causal_detected_weight1 <- sum(ctwas_genes_weight1 %in% true_genes_combined)
#load cTWAS results for weight2
ctwas_res <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR_weight2.susieIrss.txt"))
ctwas_gene_res <- ctwas_res[ctwas_res$type!="SNP",]
#number of genes with cTWAS PIP > threshold in weight1
ctwas_genes_weight2 <- ctwas_gene_res$id[ctwas_gene_res$susie_pip > PIP_threshold]
n_ctwas_genes_weight2 <- length(ctwas_genes_weight2)
n_causal_detected_weight2 <- sum(ctwas_genes_weight2 %in% true_genes_combined)
#load cTWAS results for weight3
ctwas_res <- data.table::fread(paste0(results_dir, runtag, "_simu", simutag, "_config", configtag, "_LDR_weight3.susieIrss.txt"))
ctwas_gene_res <- ctwas_res[ctwas_res$type!="SNP",]
#number of genes with cTWAS PIP > threshold in weight3
ctwas_genes_weight3 <- ctwas_gene_res$id[ctwas_gene_res$susie_pip > PIP_threshold]
n_ctwas_genes_weight3 <- length(ctwas_genes_weight3)
n_causal_detected_weight3 <- sum(ctwas_genes_weight3 %in% true_genes_combined)
#combined analysis
ctwas_genes_combined <- unique(c(ctwas_genes_weight1, ctwas_genes_weight2, ctwas_genes_weight3))
n_ctwas_genes_combined <- length(ctwas_genes_combined)
n_causal_detected_combined <- sum(ctwas_genes_combined %in% true_genes_combined)
results_current <- data.frame(simutag=as.character(simutag),
n_causal_combined=as.integer(n_causal_combined),
n_detected_weight1=as.integer(n_ctwas_genes_weight1),
n_detected_in_causal_weight1=as.integer(n_causal_detected_weight1),
n_detected_weight2=as.integer(n_ctwas_genes_weight2),
n_detected_in_causal_weight2=as.integer(n_causal_detected_weight2),
n_detected_weight3=as.integer(n_ctwas_genes_weight3),
n_detected_in_causal_weight3=as.integer(n_causal_detected_weight3),
n_detected_combined=as.integer(n_ctwas_genes_combined),
n_detected_in_causal_combined=as.integer(n_causal_detected_combined))
results_df <- rbind(results_df, results_current)
}
#results using weight1
results_df[,c("simutag", colnames(results_df)[grep("weight1", colnames(results_df))])] simutag n_detected_weight1 n_detected_in_causal_weight1
1 4-1 2 0
2 4-2 0 0
3 4-3 2 0
4 4-4 2 0
5 4-5 0 0#mean percent causal using PIP > 0.8 for weight1
sum(results_df$n_detected_in_causal_weight1)/sum(results_df$n_detected_weight1)[1] 0#results using weight2
results_df[,c("simutag", colnames(results_df)[grep("weight2", colnames(results_df))])] simutag n_detected_weight2 n_detected_in_causal_weight2
1 4-1 5 0
2 4-2 0 0
3 4-3 0 0
4 4-4 6 0
5 4-5 3 0#mean percent causal using PIP > 0.8 for weight1
sum(results_df$n_detected_in_causal_weight2)/sum(results_df$n_detected_weight2)[1] 0#results using weight3
results_df[,c("simutag", colnames(results_df)[grep("weight3", colnames(results_df))])] simutag n_detected_weight3 n_detected_in_causal_weight3
1 4-1 0 0
2 4-2 0 0
3 4-3 2 0
4 4-4 5 0
5 4-5 3 0#mean percent causal using PIP > 0.8 for weight3
sum(results_df$n_detected_in_causal_weight3)/sum(results_df$n_detected_weight3)[1] 0#results using combined analysis
results_df[,c("simutag", colnames(results_df)[grep("combined", colnames(results_df))])] simutag n_causal_combined n_detected_combined n_detected_in_causal_combined
1 4-1 0 6 0
2 4-2 0 0 0
3 4-3 0 2 0
4 4-4 0 13 0
5 4-5 0 5 0#mean percent causal using PIP > 0.8 for weight3
sum(results_df$n_detected_in_causal_combined)/sum(results_df$n_detected_combined)[1] 0#store results for figure
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight1",
count=results_df$n_detected_weight1-results_df$n_detected_in_causal_weight1,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight1",
count=results_df$n_detected_in_causal_weight1,
ifcausal=T))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight2",
count=results_df$n_detected_weight2-results_df$n_detected_in_causal_weight2,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight2",
count=results_df$n_detected_in_causal_weight2,
ifcausal=T))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight3",
count=results_df$n_detected_weight3-results_df$n_detected_in_causal_weight3,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Weight3",
count=results_df$n_detected_in_causal_weight3,
ifcausal=T))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Union",
count=results_df$n_detected_combined-results_df$n_detected_in_causal_combined,
ifcausal=F))
plot_df <- rbind(plot_df,
data.frame(simutag=results_df$simutag,
method="cTWAS Union",
count=results_df$n_detected_in_causal_combined,
ifcausal=T))Figure
For the cTWAS analysis, each tissue was analyzed individually and the results were combined.
plot_df$method <- factor(plot_df$method, levels=c("TWAS", "cTWAS G+T", "cTWAS G", "cTWAS G - Ind Var", "cTWAS Union", "cTWAS Weight1", "cTWAS Weight2", "cTWAS Weight3"))
library(ggpubr)
colset = c("#ebebeb", "#fb8072")
ggbarplot(plot_df,
x = "method",
y = "count",
add = "mean_se",
fill = "ifcausal",
legend = "none",
ylab="Count",
xlab="",
palette = colset) + grids(linetype = "dashed") + theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1))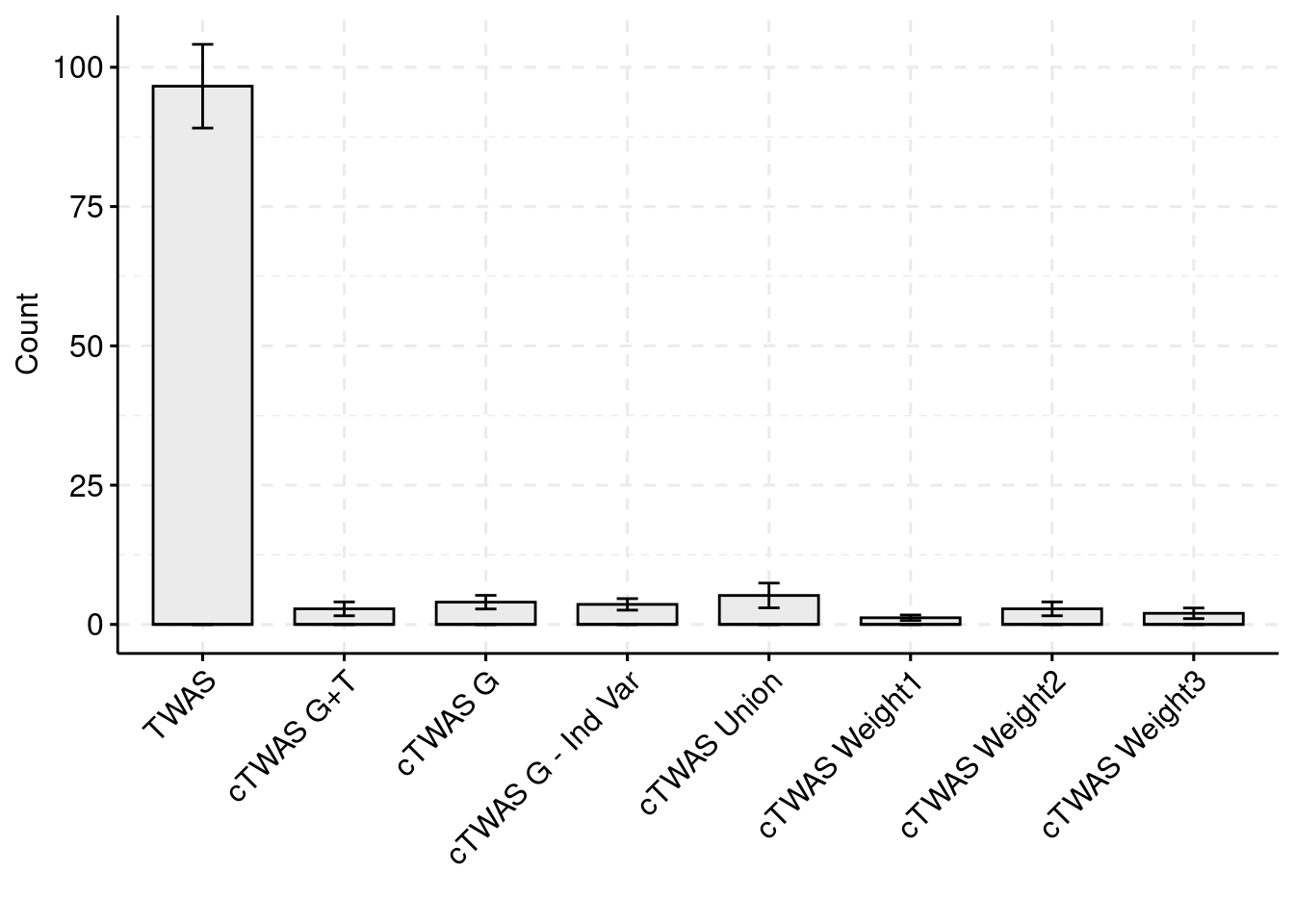
| Version | Author | Date |
|---|---|---|
| 9ed1bc7 | sq-96 | 2023-10-05 |
plot_df_2 <- plot_df
plot_df_2$method %in% c("TWAS", "cTWAS Union", "cTWAS Weight1", "cTWAS Weight2", "cTWAS Weight3") [1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[13] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE
[25] TRUE TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
[37] FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[49] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[61] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[73] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUEplot_df_2$ifcausal <- as.character(plot_df_2$ifcausal+1)
plot_df_2$count[plot_df_2$method=="cTWAS G" & plot_df_2$ifcausal=="2"] <- plot_df_2$count[plot_df_2$method=="cTWAS G" & plot_df_2$ifcausal=="2"] - plot_df_2$count[plot_df_2$method=="cTWAS G+T" & plot_df_2$ifcausal=="2"]
plot_df_2 <- rbind(plot_df_2, data.frame(simutag=simutags,
method="cTWAS G",
count=plot_df_2$count[plot_df_2$method=="cTWAS G+T" & plot_df_2$ifcausal=="2"],
ifcausal="3"))
plot_df_3 <- plot_df_2
plot_df_2 <- plot_df_2[plot_df_2$method %in% c("TWAS", "cTWAS Union", "cTWAS Weight1", "cTWAS Weight2", "cTWAS Weight3"),]
plot_df_2$method <- droplevels(plot_df_2$method)
levels(plot_df_2$method) <- c("TWAS", "cTWAS Union", "cTWAS Primary", "cTWAS Secondary", "cTWAS Null")
plot_df_2$ifcausal[plot_df_2$ifcausal=="1"] <- "FP"
plot_df_2$ifcausal[plot_df_2$ifcausal=="2"] <- "TP"
colset = c("#ebebeb", "#fb8072", "#8dd3c7")
ggbarplot(plot_df_2,
x = "method",
y = "count",
add = "mean_se",
fill = "ifcausal",
legend = "right",
ylab="Count",
xlab="",
palette = colset) + grids(linetype = "dashed") + theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1)) + guides(fill=guide_legend(title=""))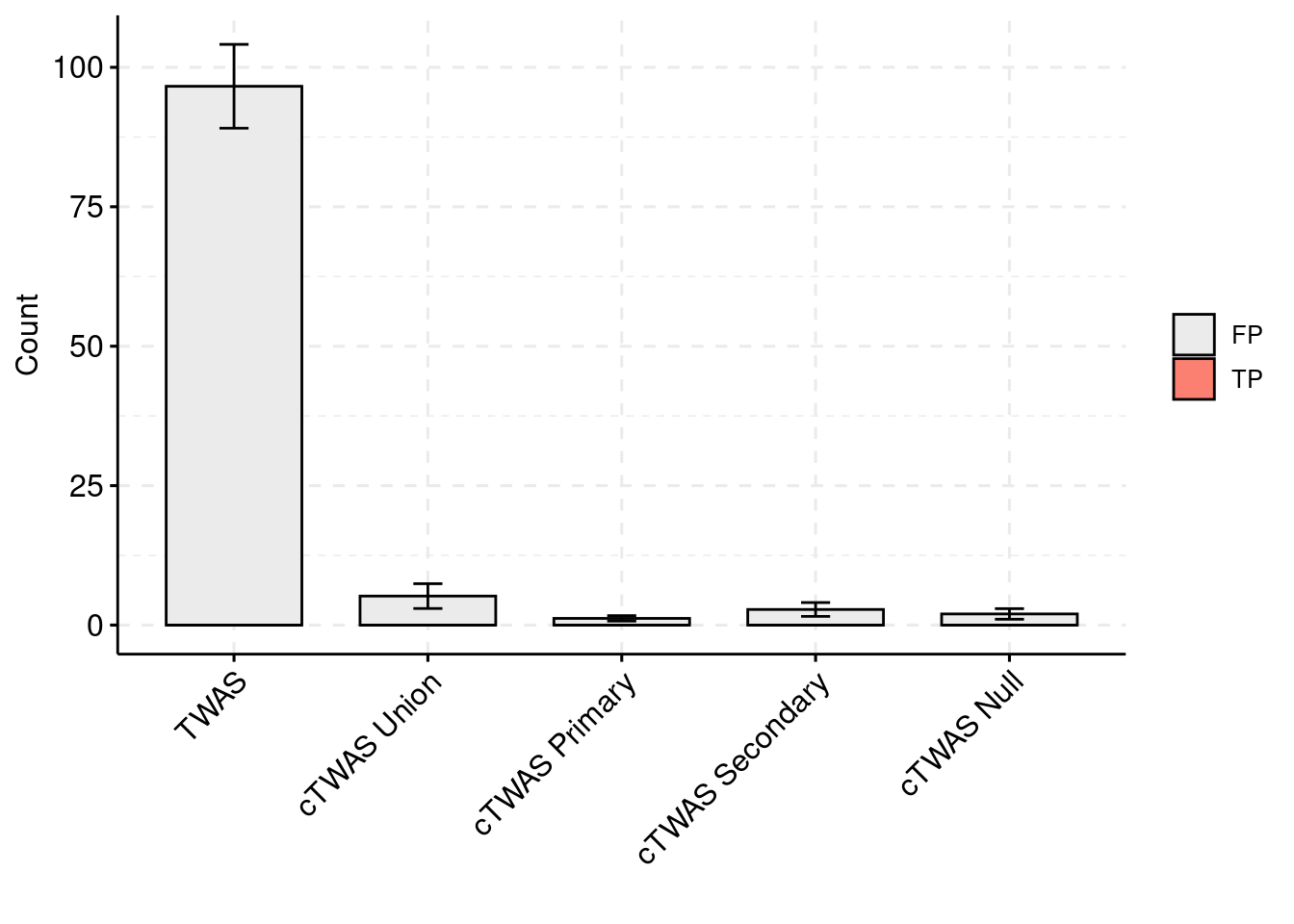
| Version | Author | Date |
|---|---|---|
| 9ed1bc7 | sq-96 | 2023-10-05 |
plot_df_3 <- plot_df_3[plot_df_3$method %in% c("cTWAS G", "cTWAS Union"),]
plot_df_3$method <- droplevels(plot_df_3$method)
levels(plot_df_3$method) <- c("cTWAS Joint", "cTWAS Union")
plot_df_3$method <- relevel(plot_df_3$method, "cTWAS Union")
plot_df_3$ifcausal[plot_df_3$ifcausal=="1"] <- "FP"
plot_df_3$ifcausal[plot_df_3$ifcausal=="2"] <- "TP"
plot_df_3$ifcausal[plot_df_3$ifcausal=="3"] <- "TP and Tissue Identified\nby Joint Analysis"
ggbarplot(plot_df_3,
x = "method",
y = "count",
add = "mean_se",
fill = "ifcausal",
legend = "right",
ylab="Count",
xlab="",
palette = colset) + grids(linetype = "dashed") + theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1)) + guides(fill=guide_legend(title=""))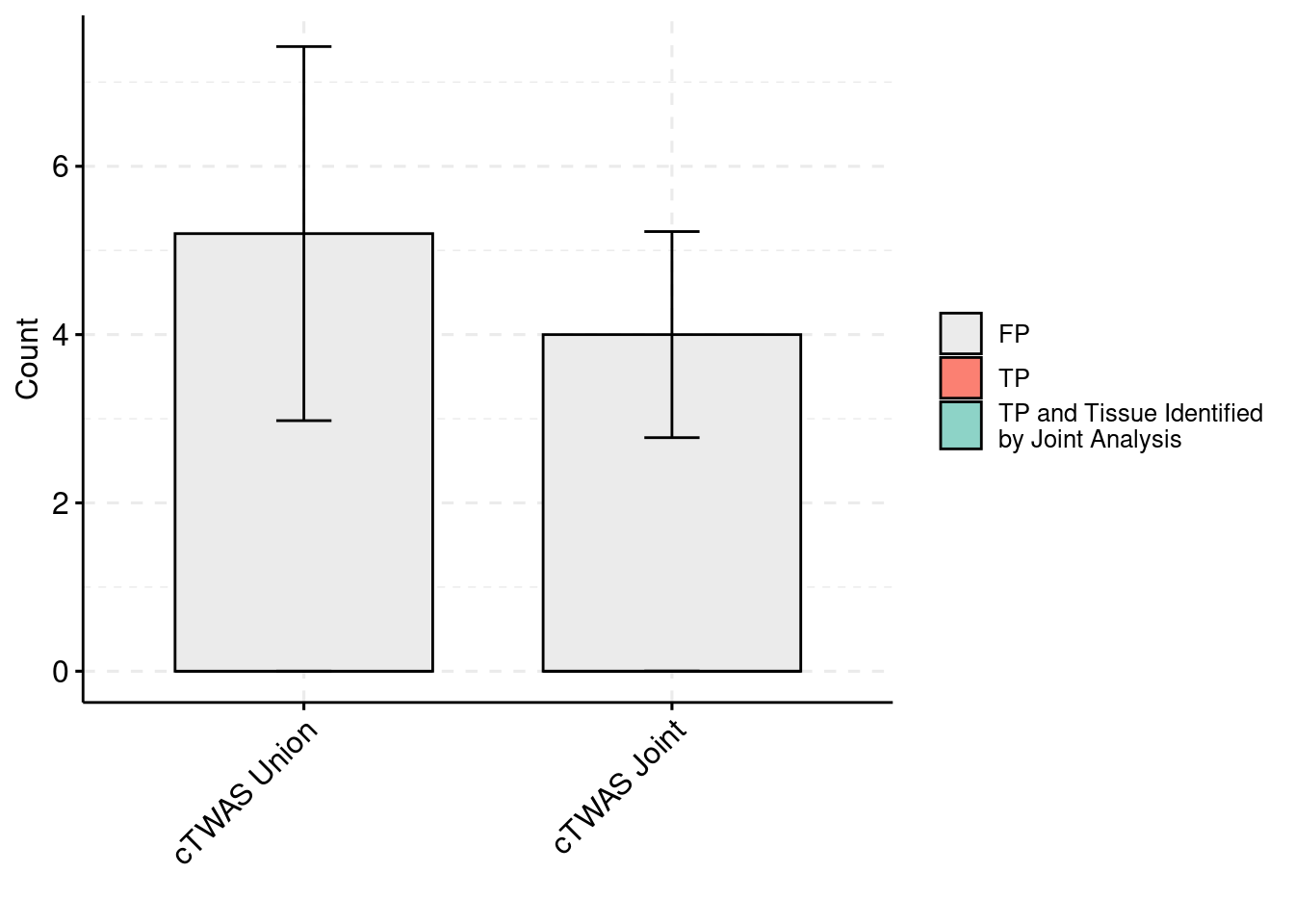
| Version | Author | Date |
|---|---|---|
| 9ed1bc7 | sq-96 | 2023-10-05 |
sessionInfo()R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggpubr_0.6.0 ggplot2_3.4.0 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] tidyselect_1.2.0 xfun_0.35 bslib_0.4.1 purrr_1.0.2
[5] carData_3.0-4 colorspace_2.0-3 vctrs_0.6.3 generics_0.1.3
[9] htmltools_0.5.4 yaml_2.3.6 utf8_1.2.2 rlang_1.1.1
[13] jquerylib_0.1.4 later_1.3.0 pillar_1.8.1 glue_1.6.2
[17] withr_2.5.0 DBI_1.1.3 lifecycle_1.0.3 stringr_1.5.0
[21] ggsignif_0.6.4 munsell_0.5.0 gtable_0.3.1 evaluate_0.19
[25] labeling_0.4.2 knitr_1.41 callr_3.7.3 fastmap_1.1.0
[29] httpuv_1.6.7 ps_1.7.2 fansi_1.0.3 highr_0.9
[33] broom_1.0.2 Rcpp_1.0.9 backports_1.2.1 promises_1.2.0.1
[37] scales_1.2.1 cachem_1.0.6 jsonlite_1.8.4 abind_1.4-5
[41] farver_2.1.0 fs_1.5.2 digest_0.6.31 stringi_1.7.8
[45] rstatix_0.7.2 processx_3.8.0 dplyr_1.0.10 getPass_0.2-2
[49] rprojroot_2.0.3 grid_4.1.0 cli_3.6.1 tools_4.1.0
[53] magrittr_2.0.3 sass_0.4.4 tibble_3.1.8 car_3.1-1
[57] tidyr_1.3.0 whisker_0.4.1 pkgconfig_2.0.3 data.table_1.14.6
[61] assertthat_0.2.1 rmarkdown_2.19 httr_1.4.4 rstudioapi_0.14
[65] R6_2.5.1 git2r_0.30.1 compiler_4.1.0