Simulation Results using three Tissues (uncorrelated)
shengqian
2023-10-13
Last updated: 2023-10-21
Checks: 5 2
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/analysis/simulation_help_functions.R | analysis/simulation_help_functions.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 3122878. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .ipynb_checkpoints/
Untracked files:
Untracked: LDL_LDLR_locus1.pdf
Untracked: LDL_TEME199_genetrack.pdf
Untracked: LDL_TEME199_locus.pdf
Untracked: Proposal plots.R
Untracked: RGS14.pdf
Untracked: RNF186.pdf
Untracked: Rplots.pdf
Untracked: SCZ_annotation.xlsx
Untracked: SLC8B1.pdf
Untracked: analysis/.ipynb_checkpoints/
Untracked: cache/
Untracked: code/.ipynb_checkpoints/
Untracked: data/.ipynb_checkpoints/
Untracked: data/FUMA_output/
Untracked: data/GO_Terms/
Untracked: data/GTEx_Analysis_v8_eQTL.tar
Untracked: data/G_list.RData
Untracked: data/IBD_ME/
Untracked: data/LDL/
Untracked: data/LDL_E_S/
Untracked: data/LDL_E_S_M/
Untracked: data/LDL_M/
Untracked: data/LDL_S/
Untracked: data/PGC3_SCZ_wave3_public.v2.tsv
Untracked: data/Predictive_Models/
Untracked: data/Supplementary Table 15 - MAGMA.xlsx
Untracked: data/Supplementary Table 20 - Prioritised Genes.xlsx
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/WhiteBlood_E/
Untracked: data/WhiteBlood_E_M/
Untracked: data/WhiteBlood_E_S_M/
Untracked: data/WhiteBlood_E_S_M_PC/
Untracked: data/WhiteBlood_M/
Untracked: data/WhiteBlood_M_compare/
Untracked: data/WhiteBlood_M_enet/
Untracked: data/cpg_annot.RData
Untracked: data/eqtl/
Untracked: data/gencode.v26.GRCh38.genes.gtf
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/gwas_sumstats/
Untracked: data/magma.genes.out
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/mqtl/
Untracked: data/notes.txt
Untracked: data/scz_2018.RDS
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/test/
Untracked: hist.pdf
Untracked: ld_pip.pdf
Untracked: submit.sh
Untracked: temp_LDR/
Untracked: test-B1.snpgwas.txt
Unstaged changes:
Deleted: analysis/Atrial_Fibrillation_Heart_Atrial_Appendage.Rmd
Deleted: analysis/Atrial_Fibrillation_Heart_Left_Ventricle.Rmd
Deleted: analysis/Autism_Brain_Amygdala.Rmd
Deleted: analysis/Autism_Brain_Anterior_cingulate_cortex_BA24.Rmd
Deleted: analysis/Autism_Brain_Caudate_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Cerebellar_Hemisphere.Rmd
Deleted: analysis/Autism_Brain_Cerebellum.Rmd
Deleted: analysis/Autism_Brain_Cortex.Rmd
Deleted: analysis/Autism_Brain_Frontal_Cortex_BA9.Rmd
Deleted: analysis/Autism_Brain_Hippocampus.Rmd
Deleted: analysis/Autism_Brain_Hypothalamus.Rmd
Deleted: analysis/Autism_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Putamen_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Spinal_cord_cervical_c-1.Rmd
Deleted: analysis/Autism_Brain_Substantia_nigra.Rmd
Deleted: analysis/BMI_Brain_Amygdala.Rmd
Deleted: analysis/BMI_Brain_Amygdala_S.Rmd
Deleted: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24.Rmd
Deleted: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24_S.Rmd
Deleted: analysis/BMI_Brain_Caudate_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Caudate_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Cerebellar_Hemisphere.Rmd
Deleted: analysis/BMI_Brain_Cerebellar_Hemisphere_S.Rmd
Deleted: analysis/BMI_Brain_Cerebellum.Rmd
Deleted: analysis/BMI_Brain_Cerebellum_S.Rmd
Deleted: analysis/BMI_Brain_Cortex.Rmd
Deleted: analysis/BMI_Brain_Cortex_S.Rmd
Deleted: analysis/BMI_Brain_Frontal_Cortex_BA9.Rmd
Deleted: analysis/BMI_Brain_Frontal_Cortex_BA9_S.Rmd
Deleted: analysis/BMI_Brain_Hippocampus.Rmd
Deleted: analysis/BMI_Brain_Hippocampus_S.Rmd
Deleted: analysis/BMI_Brain_Hypothalamus.Rmd
Deleted: analysis/BMI_Brain_Hypothalamus_S.Rmd
Deleted: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Putamen_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Putamen_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Spinal_cord_cervical_c-1.Rmd
Deleted: analysis/BMI_Brain_Spinal_cord_cervical_c-1_S.Rmd
Deleted: analysis/BMI_Brain_Substantia_nigra.Rmd
Deleted: analysis/BMI_Brain_Substantia_nigra_S.Rmd
Deleted: analysis/BMI_S_results.Rmd
Deleted: analysis/Glucose_Adipose_Subcutaneous.Rmd
Deleted: analysis/Glucose_Adipose_Visceral_Omentum.Rmd
Modified: analysis/sim_uncor_3_d_m.Rmd
Modified: analysis/simulation_3tissues_correlated_merge.Rmd
Modified: analysis/simulation_3tissues_uncorrelated_merge.Rmd
Modified: analysis/simulation_help_functions.R
Deleted: code/WhiteBlood_M_ener_out/WhiteBlood_WholeBlood.out
Deleted: code/White_Blood_M_out/White_Blood_BreastMammary.err
Deleted: code/White_Blood_M_out/White_Blood_BreastMammary.out
Deleted: code/White_Blood_M_out/White_Blood_ColonTransverse.err
Deleted: code/White_Blood_M_out/White_Blood_ColonTransverse.out
Deleted: code/White_Blood_M_out/White_Blood_KidneyCortex.err
Deleted: code/White_Blood_M_out/White_Blood_KidneyCortex.out
Deleted: code/White_Blood_M_out/White_Blood_Lung.err
Deleted: code/White_Blood_M_out/White_Blood_Lung.out
Deleted: code/White_Blood_M_out/White_Blood_MuscleSkeletal.err
Deleted: code/White_Blood_M_out/White_Blood_MuscleSkeletal.out
Deleted: code/White_Blood_M_out/White_Blood_Ovary.err
Deleted: code/White_Blood_M_out/White_Blood_Ovary.out
Deleted: code/White_Blood_M_out/White_Blood_Prostate.err
Deleted: code/White_Blood_M_out/White_Blood_Prostate.out
Deleted: code/White_Blood_M_out/White_Blood_Testis.err
Deleted: code/White_Blood_M_out/White_Blood_Testis.out
Deleted: code/White_Blood_M_out/White_Blood_WholeBlood.err
Deleted: code/White_Blood_M_out/White_Blood_WholeBlood.out
Deleted: code/run_IBD_ctwas_rss_LDR_ME.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/sim_uncor_3_d_m.Rmd) and HTML (docs/sim_uncor_3_d_m.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 3122878 | sq-96 | 2023-10-20 | update |
| html | c5c7a7f | sq-96 | 2023-10-20 | update |
| html | 5583c52 | sq-96 | 2023-10-20 | update |
| Rmd | c10305c | sq-96 | 2023-10-20 | update |
| html | c10305c | sq-96 | 2023-10-20 | update |
| Rmd | f296597 | sq-96 | 2023-10-20 | update |
| html | f296597 | sq-96 | 2023-10-20 | update |
source("/project2/xinhe/shengqian/cTWAS/cTWAS_analysis/analysis/simulation_help_functions.R")Simulation 1: Three causal tissues with equal high PVE (3% each)
Separate effect size parameters
30% PVE and 2.5e-4 prior inclusion for SNPs, 3% PVE and 0.009 prior inclusion for Liver expression, 3% PVE and 0.009 prior inclusion for Lung expression, 3% PVE and 0.009 prior inclusion for Brain Hippocampus expression. For the cTWAS analysis, each tissue had its own prior inclusion parameter and effect size parameter.
#results using PIP threshold (gene+tissue)
results_df[,c("simutag", "n_causal", "n_detected_pip", "n_detected_pip_in_causal")] simutag n_causal n_detected_pip n_detected_pip_in_causal
1 1-1 211 18 15
2 1-2 228 33 32
3 1-3 199 19 19
4 1-4 202 15 14
5 1-5 230 45 35#mean percent causal using PIP > 0.8
sum(results_df$n_detected_pip_in_causal)/sum(results_df$n_detected_pip)[1] 0.8846154#results using combined PIP threshold
results_df[,c("simutag", "n_causal_combined", "n_detected_comb_pip", "n_detected_comb_pip_in_causal")] simutag n_causal_combined n_detected_comb_pip n_detected_comb_pip_in_causal
1 1-1 211 31 26
2 1-2 228 53 50
3 1-3 199 29 28
4 1-4 201 39 34
5 1-5 229 72 57#mean percent causal using combined PIP > 0.8
sum(results_df$n_detected_comb_pip_in_causal)/sum(results_df$n_detected_comb_pip)[1] 0.8705357#prior inclusion and mean prior inclusion
results_df[,c(which(colnames(results_df)=="simutag"), setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df))))] simutag prior_snp prior_weight1 prior_weight2 prior_weight3
1 1-1 0.0002425920 0.010177436 0.008623747 0.004769606
2 1-2 0.0002466862 0.010292635 0.003108254 0.009989375
3 1-3 0.0002626968 0.005504924 0.009932813 0.006229771
4 1-4 0.0002604598 0.002595350 0.011445972 0.008293892
5 1-5 0.0002372624 0.005251018 0.019663392 0.009663112colMeans(results_df[,setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df)))]) prior_snp prior_weight1 prior_weight2 prior_weight3
0.0002499394 0.0067642725 0.0105548357 0.0077891513 #prior variance and mean prior variance
results_df[,c(which(colnames(results_df)=="simutag"), grep("prior_var", names(results_df)))] simutag prior_var_snp prior_var_weight1 prior_var_weight2 prior_var_weight3
1 1-1 9.038498 3.856506 14.351212 32.70334
2 1-2 8.226705 15.565359 35.320799 18.97877
3 1-3 8.540242 28.372561 6.987738 26.73232
4 1-4 8.742224 15.309434 10.017471 26.50675
5 1-5 8.260003 30.294016 13.022285 23.62614colMeans(results_df[,grep("prior_var", names(results_df))]) prior_var_snp prior_var_weight1 prior_var_weight2 prior_var_weight3
8.561535 18.679575 15.939901 25.709463 #PVE and mean PVE
results_df[,c(which(colnames(results_df)=="simutag"), grep("pve", names(results_df)))] simutag pve_snp pve_weight1 pve_weight2 pve_weight3
1 1-1 0.2581101 0.006449976 0.02688094 0.02759495
2 1-2 0.2388928 0.026327607 0.02384552 0.03353988
3 1-3 0.2640931 0.025667024 0.01507540 0.02946211
4 1-4 0.2680371 0.006529511 0.02490409 0.03889286
5 1-5 0.2306967 0.026141228 0.05561673 0.04038916colMeans(results_df[,grep("pve", names(results_df))]) pve_snp pve_weight1 pve_weight2 pve_weight3
0.25196596 0.01822307 0.02926453 0.03397579 #TWAS results
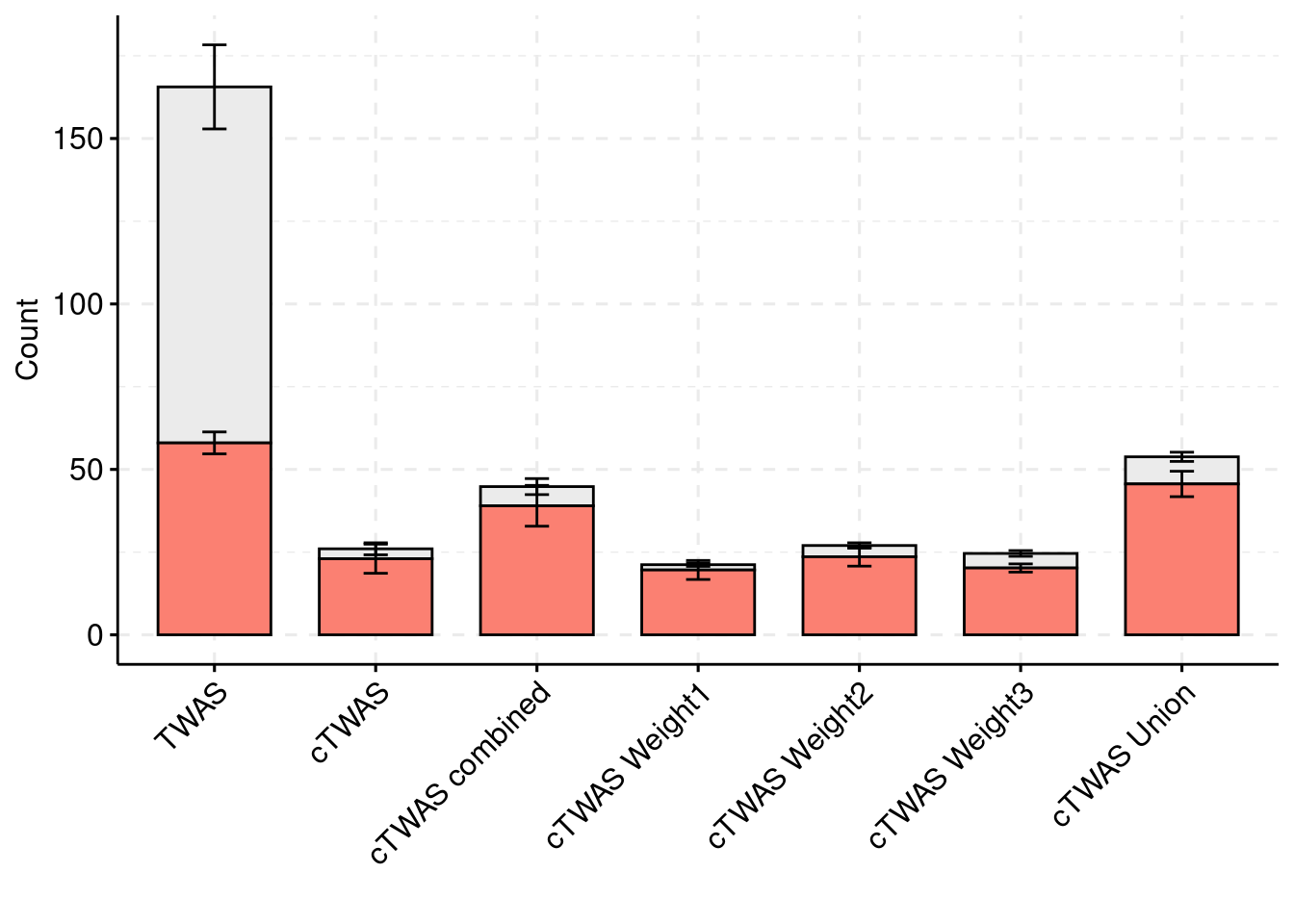
results_df[,c(which(colnames(results_df)=="simutag"), grep("twas", names(results_df)))] simutag n_detected_twas n_detected_twas_in_causal n_detected_comb_twas
1 1-1 247 55 160
2 1-2 326 65 200
3 1-3 327 59 198
4 1-4 214 47 124
5 1-5 230 63 146
n_detected_comb_twas_in_causal
1 55
2 66
3 59
4 47
5 63sum(results_df$n_detected_comb_twas_in_causal)/sum(results_df$n_detected_comb_twas)[1] 0.3502415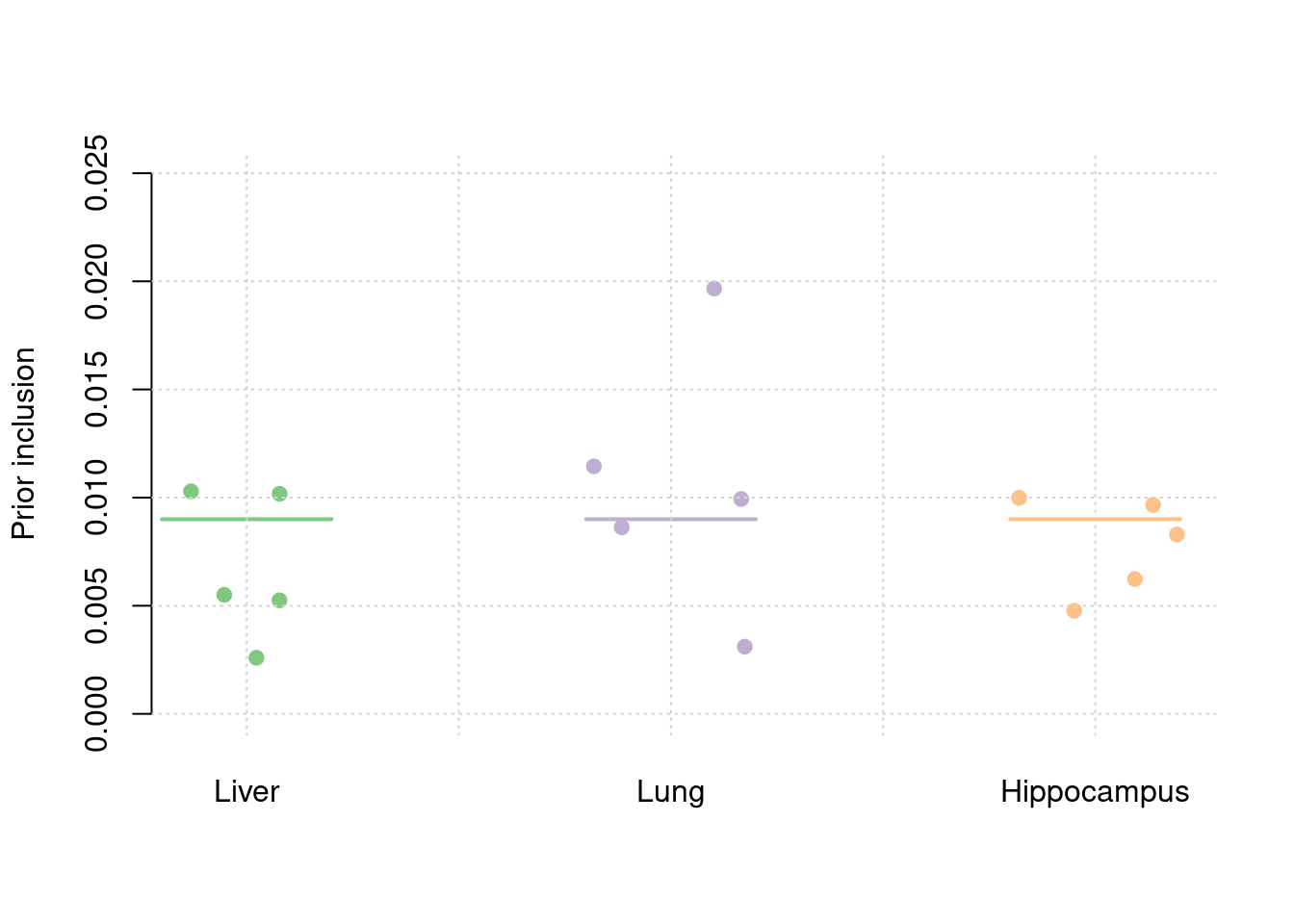
| Version | Author | Date |
|---|---|---|
| f296597 | sq-96 | 2023-10-20 |
| Version | Author | Date |
|---|---|---|
| f296597 | sq-96 | 2023-10-20 |
| Version | Author | Date |
|---|---|---|
| f296597 | sq-96 | 2023-10-20 |
Individual tissue analyses
#results using weight1
results_df[,c("simutag", colnames(results_df)[grep("weight1", colnames(results_df))])] simutag n_detected_weight1 n_detected_in_causal_weight1
1 1-1 21 18
2 1-2 32 30
3 1-3 21 21
4 1-4 15 14
5 1-5 17 15#mean percent causal using PIP > 0.8 for weight1
sum(results_df$n_detected_in_causal_weight1)/sum(results_df$n_detected_weight1)[1] 0.9245283#results using weight2
results_df[,c("simutag", colnames(results_df)[grep("weight2", colnames(results_df))])] simutag n_detected_weight2 n_detected_in_causal_weight2
1 1-1 27 22
2 1-2 28 26
3 1-3 20 16
4 1-4 22 21
5 1-5 38 33#mean percent causal using PIP > 0.8 for weight1
sum(results_df$n_detected_in_causal_weight2)/sum(results_df$n_detected_weight2)[1] 0.8740741#results using weight3
results_df[,c("simutag", colnames(results_df)[grep("weight3", colnames(results_df))])] simutag n_detected_weight3 n_detected_in_causal_weight3
1 1-1 22 19
2 1-2 25 20
3 1-3 21 19
4 1-4 23 18
5 1-5 32 25#mean percent causal using PIP > 0.8 for weight3
sum(results_df$n_detected_in_causal_weight3)/sum(results_df$n_detected_weight3)[1] 0.8211382#results using combined analysis
results_df[,c("simutag", colnames(results_df)[grep("combined", colnames(results_df))])] simutag n_causal_combined n_detected_combined n_detected_in_causal_combined
1 1-1 211 53 44
2 1-2 228 59 51
3 1-3 199 47 41
4 1-4 201 40 35
5 1-5 229 70 57#mean percent causal using PIP > 0.8 for weight3
sum(results_df$n_detected_in_causal_combined)/sum(results_df$n_detected_combined)[1] 0.8475836Simulation 2: Three causal tissues with equal low PVE (1% each)
Separate effect size parameters
30% PVE and 2.5e-4 prior inclusion for SNPs, 1% PVE and 0.003 prior inclusion for Liver expression, 1% PVE and 0.003 prior inclusion for Lung expression, 1% PVE and 0.003 prior inclusion for Brain Hippocampus expression. Each tissue had its own prior inclusion parameter and effect size parameter.
results_dir <- "/project2/xinhe/shengqian/cTWAS/cTWAS_simulation/simulation_uncorrelated_drop_merge/"
runtag = "ukb-s80.45-3_uncorr"
configtag <- 2
simutags <- paste(2, 1:5, sep = "-")
thin <- 0.1
sample_size <- 45000
PIP_threshold <- 0.8results_df <- get_sim_joint_res(results_dir,runtag,configtag,simutags,thin,sample_size,PIP_threshold)#results using PIP threshold (gene+tissue)
results_df[,c("simutag", "n_causal", "n_detected_pip", "n_detected_pip_in_causal")] simutag n_causal n_detected_pip n_detected_pip_in_causal
1 2-1 66 10 10
2 2-2 80 12 11
3 2-3 60 9 9
4 2-4 70 7 6
5 2-5 67 2 0#mean percent causal using PIP > 0.8
sum(results_df$n_detected_pip_in_causal)/sum(results_df$n_detected_pip)[1] 0.9#results using combined PIP threshold
results_df[,c("simutag", "n_causal_combined", "n_detected_comb_pip", "n_detected_comb_pip_in_causal")] simutag n_causal_combined n_detected_comb_pip n_detected_comb_pip_in_causal
1 2-1 66 16 14
2 2-2 80 19 17
3 2-3 60 14 13
4 2-4 70 11 9
5 2-5 67 10 6#mean percent causal using combined PIP > 0.8
sum(results_df$n_detected_comb_pip_in_causal)/sum(results_df$n_detected_comb_pip)[1] 0.8428571#prior inclusion and mean prior inclusion
results_df[,c(which(colnames(results_df)=="simutag"), setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df))))] simutag prior_snp prior_weight1 prior_weight2 prior_weight3
1 2-1 0.0002315651 0.001568621 0.002224819 0.004115142
2 2-2 0.0002800566 0.003182041 0.004210946 0.007715860
3 2-3 0.0002814913 0.007796100 0.002747631 0.005283211
4 2-4 0.0002955710 0.002673367 0.010048005 0.003678914
5 2-5 0.0002608473 0.004279611 0.003968238 0.002141939colMeans(results_df[,setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df)))]) prior_snp prior_weight1 prior_weight2 prior_weight3
0.0002699062 0.0038999481 0.0046399278 0.0045870132 #prior variance and mean prior variance
results_df[,c(which(colnames(results_df)=="simutag"), grep("prior_var", names(results_df)))] simutag prior_var_snp prior_var_weight1 prior_var_weight2 prior_var_weight3
1 2-1 8.767998 68.34907 28.857947 20.589987
2 2-2 7.609580 38.69702 7.795359 19.540985
3 2-3 7.387488 11.73957 21.807436 12.946441
4 2-4 7.829719 21.89263 7.033374 9.215843
5 2-5 7.889612 11.77339 21.272685 13.379534colMeans(results_df[,grep("prior_var", names(results_df))]) prior_var_snp prior_var_weight1 prior_var_weight2 prior_var_weight3
7.896879 30.490337 17.353360 15.134558 #PVE and mean PVE
results_df[,c(which(colnames(results_df)=="simutag"), grep("pve", names(results_df)))] simutag pve_snp pve_weight1 pve_weight2 pve_weight3
1 2-1 0.2390043 0.017618804 0.013945043 0.014989805
2 2-2 0.2508642 0.020235268 0.007129772 0.026673860
3 2-3 0.2447901 0.015040262 0.013014363 0.012100505
4 2-4 0.2724208 0.009617943 0.015349823 0.005998047
5 2-5 0.2422558 0.008280022 0.018334952 0.005069945colMeans(results_df[,grep("pve", names(results_df))]) pve_snp pve_weight1 pve_weight2 pve_weight3
0.24986705 0.01415846 0.01355479 0.01296643 #TWAS results
results_df[,c(which(colnames(results_df)=="simutag"), grep("twas", names(results_df)))] simutag n_detected_twas n_detected_twas_in_causal n_detected_comb_twas
1 2-1 160 20 103
2 2-2 171 24 107
3 2-3 195 24 126
4 2-4 124 20 81
5 2-5 126 14 81
n_detected_comb_twas_in_causal
1 20
2 24
3 24
4 20
5 14sum(results_df$n_detected_comb_twas_in_causal)/sum(results_df$n_detected_comb_twas)[1] 0.2048193y1 <- results_df$prior_weight1
y2 <- results_df$prior_weight2
y3 <- results_df$prior_weight3
truth <- rbind(c(1,0.003),c(2,0.003),c(3,0.003))
est <- rbind(cbind(1,y1),cbind(2,y2),cbind(3,y3))
plot_par(truth,est,xlabels = c("Liver","Lung","Hippocampus"),ylim=c(0,0.025),ylab="Prior inclusion")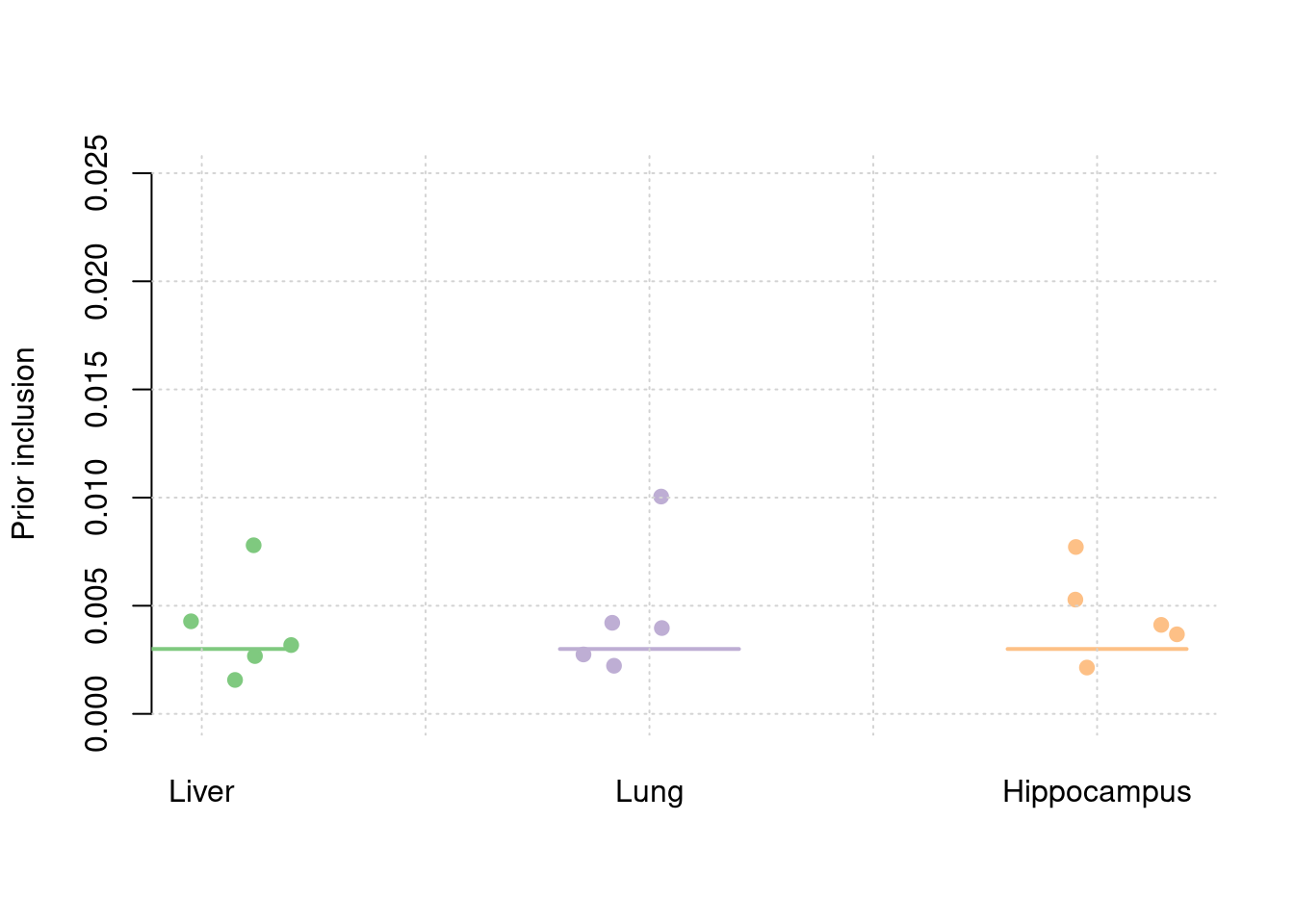
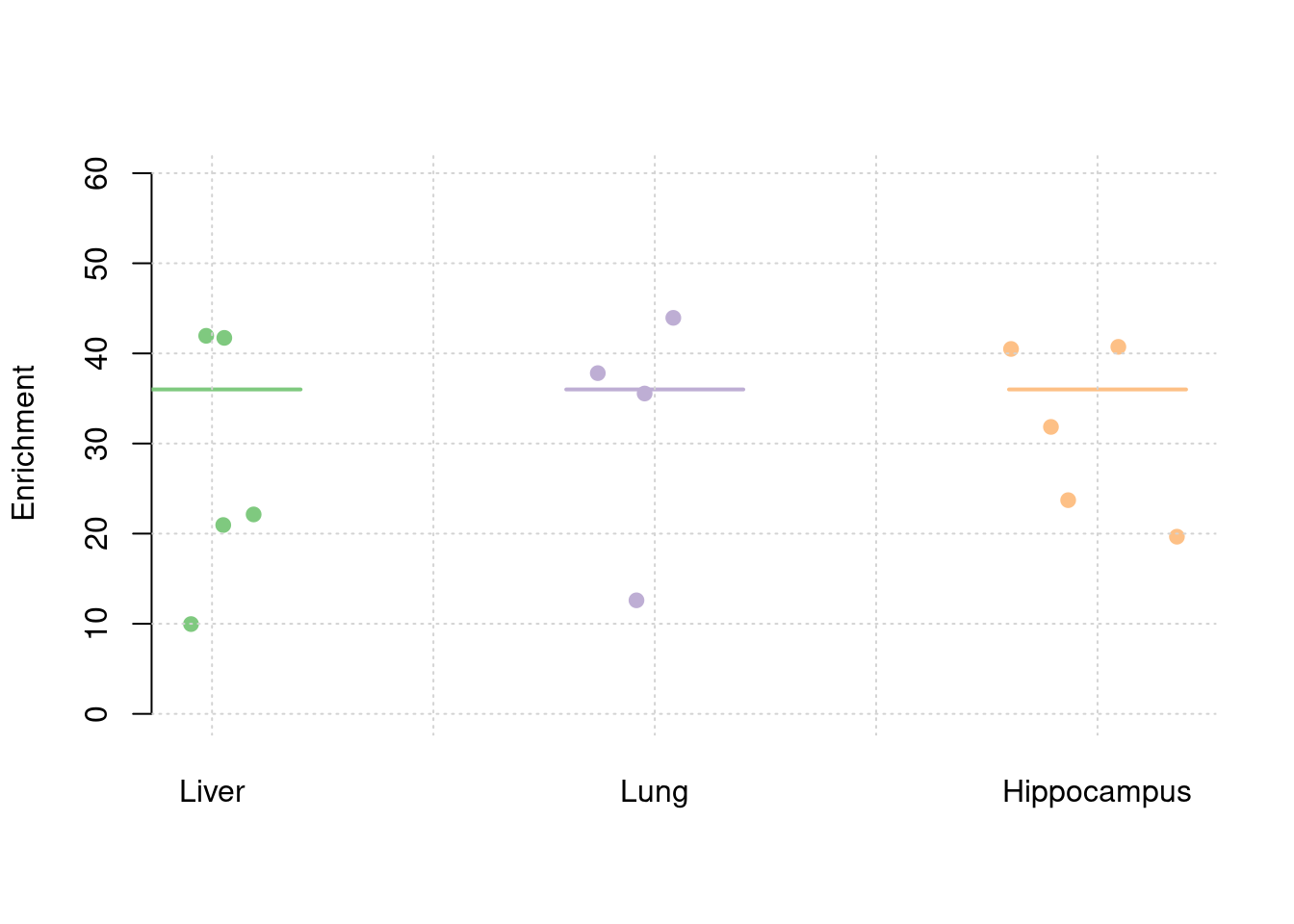
y1 <- results_df$prior_weight1/results_df$prior_snp
y2 <- results_df$prior_weight2/results_df$prior_snp
y3 <- results_df$prior_weight3/results_df$prior_snp
truth <- rbind(c(1,12),c(2,12),c(3,12))
est <- rbind(cbind(1,y1),cbind(2,y2),cbind(3,y3))
plot_par(truth,est,xlabels = c("Liver","Lung","Hippocampus"),ylim=c(0,60),ylab="Enrichment")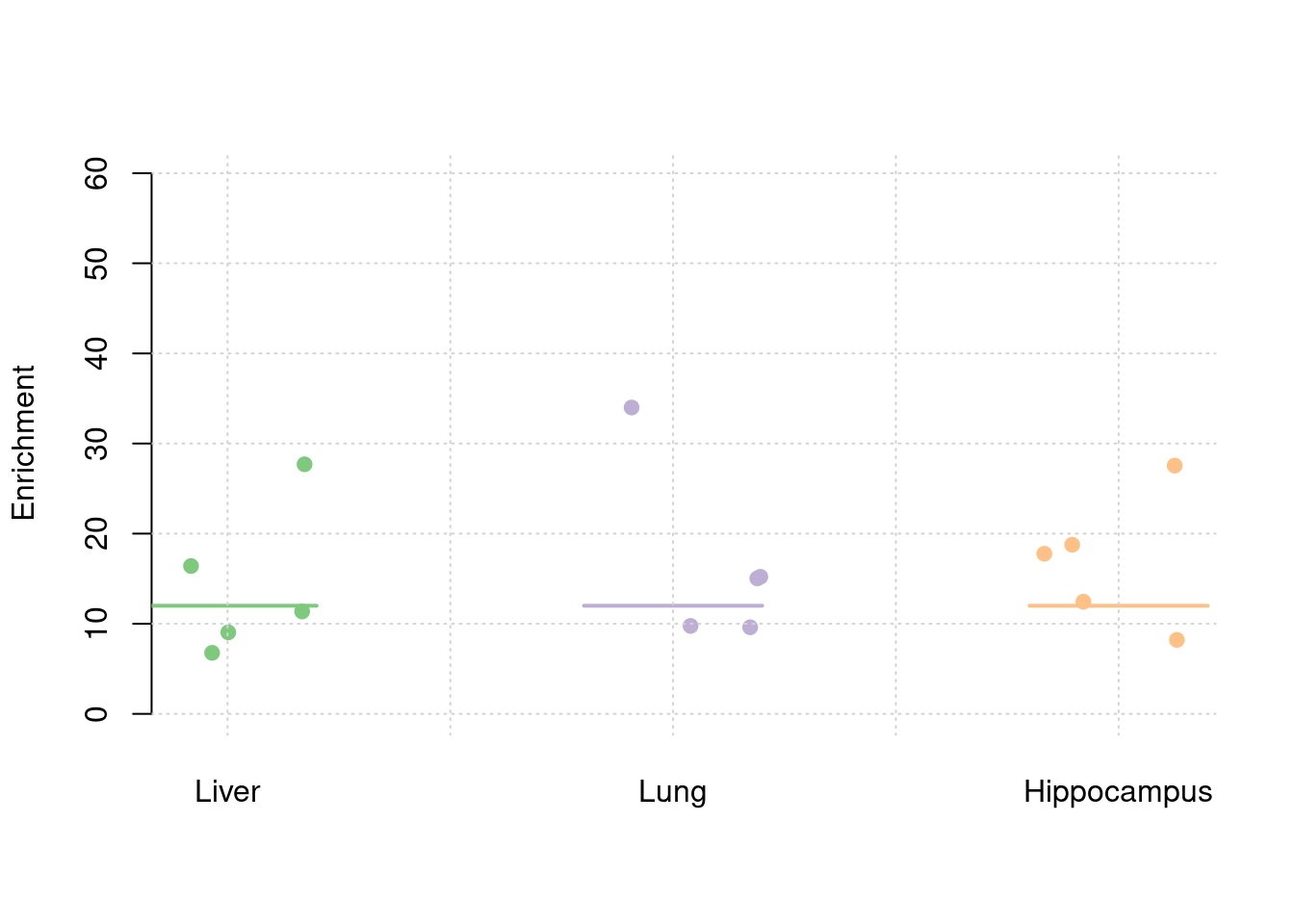
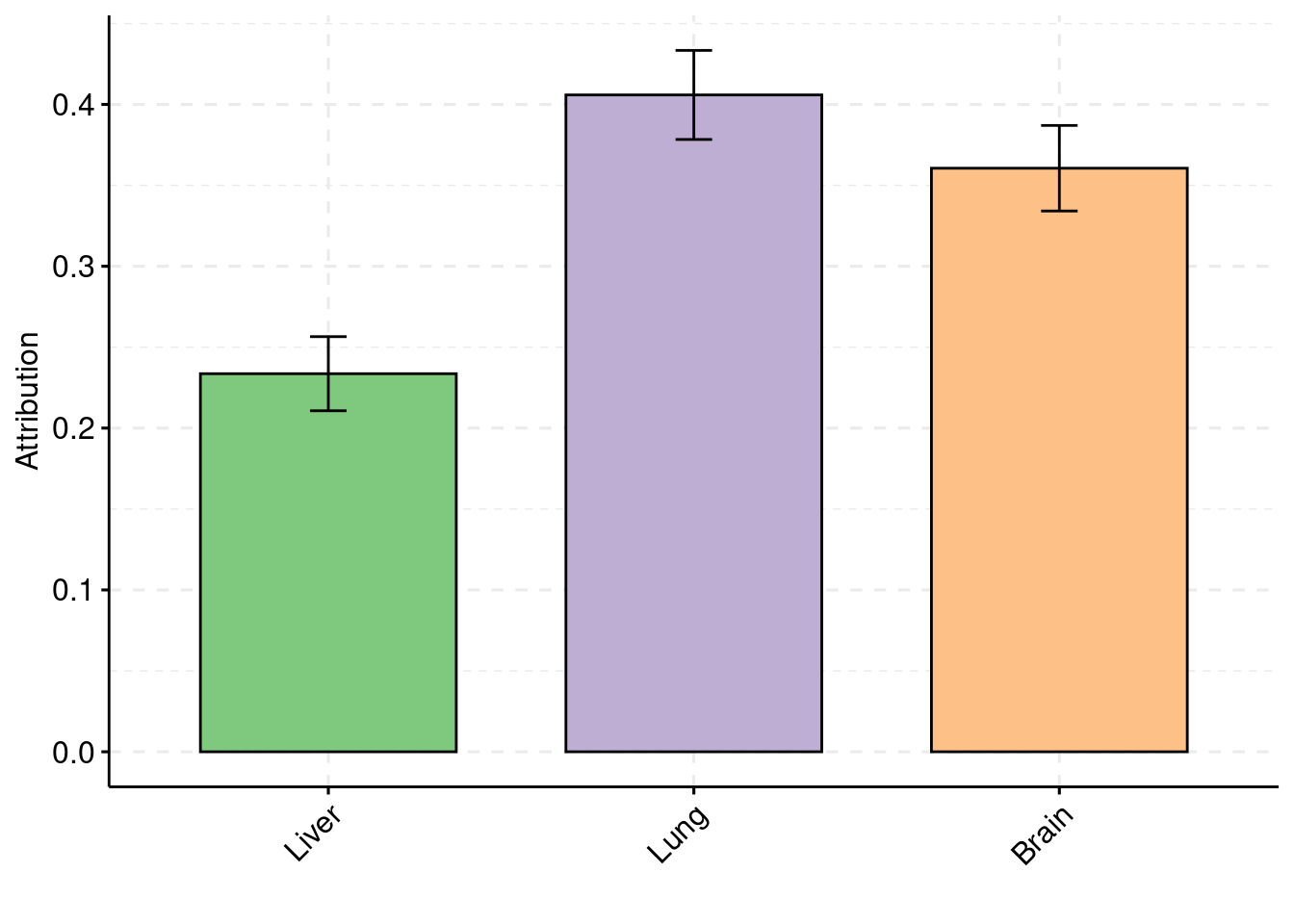
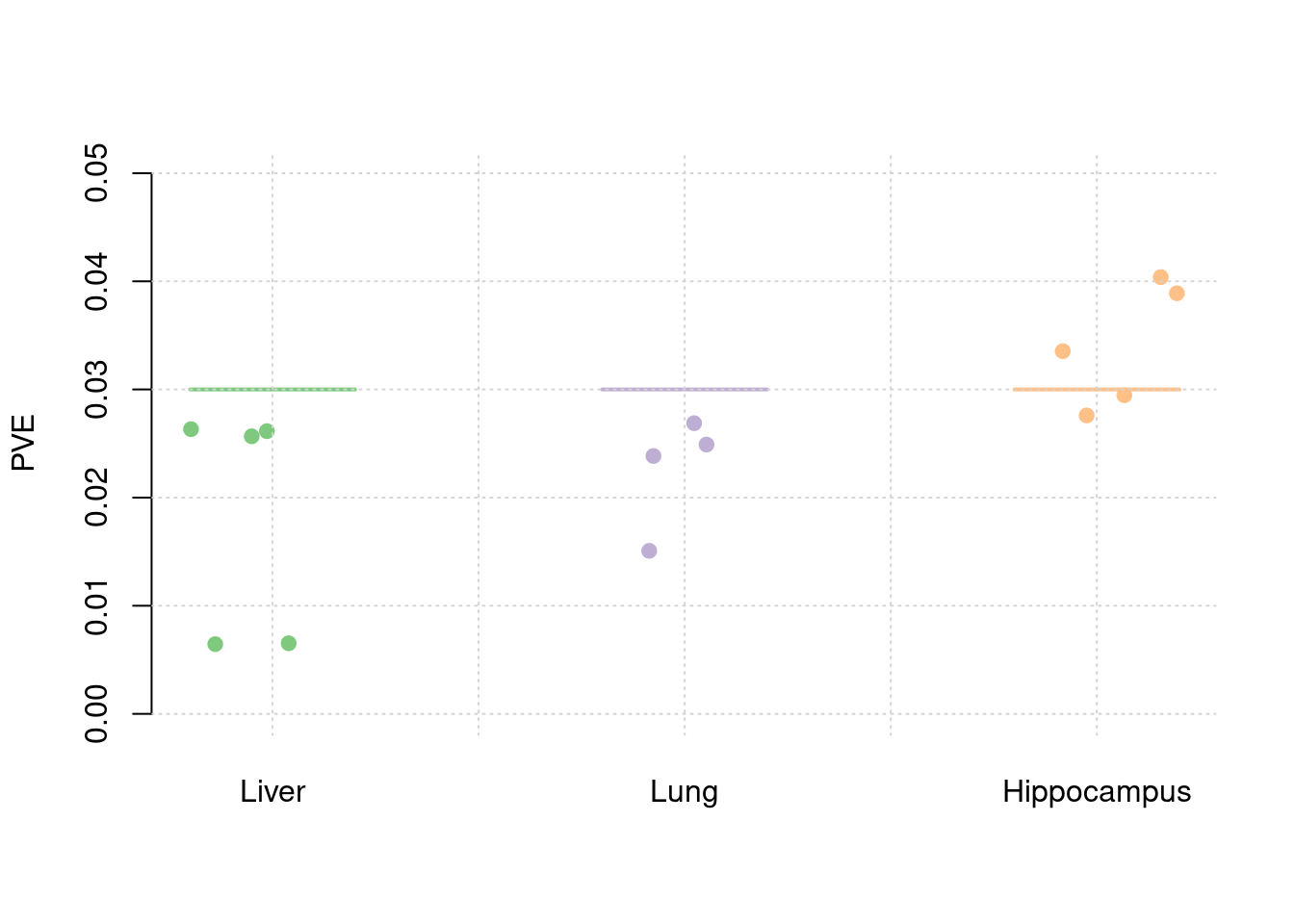
y1 <- results_df$pve_weight1
y2 <- results_df$pve_weight2
y3 <- results_df$pve_weight3
truth <- rbind(c(1,0.01),c(2,0.01),c(3,0.01))
est <- rbind(cbind(1,y1),cbind(2,y2),cbind(3,y3))
plot_par(truth,est,xlabels = c("Liver","Lung","Hippocampus"),ylim=c(0,0.05),ylab="PVE")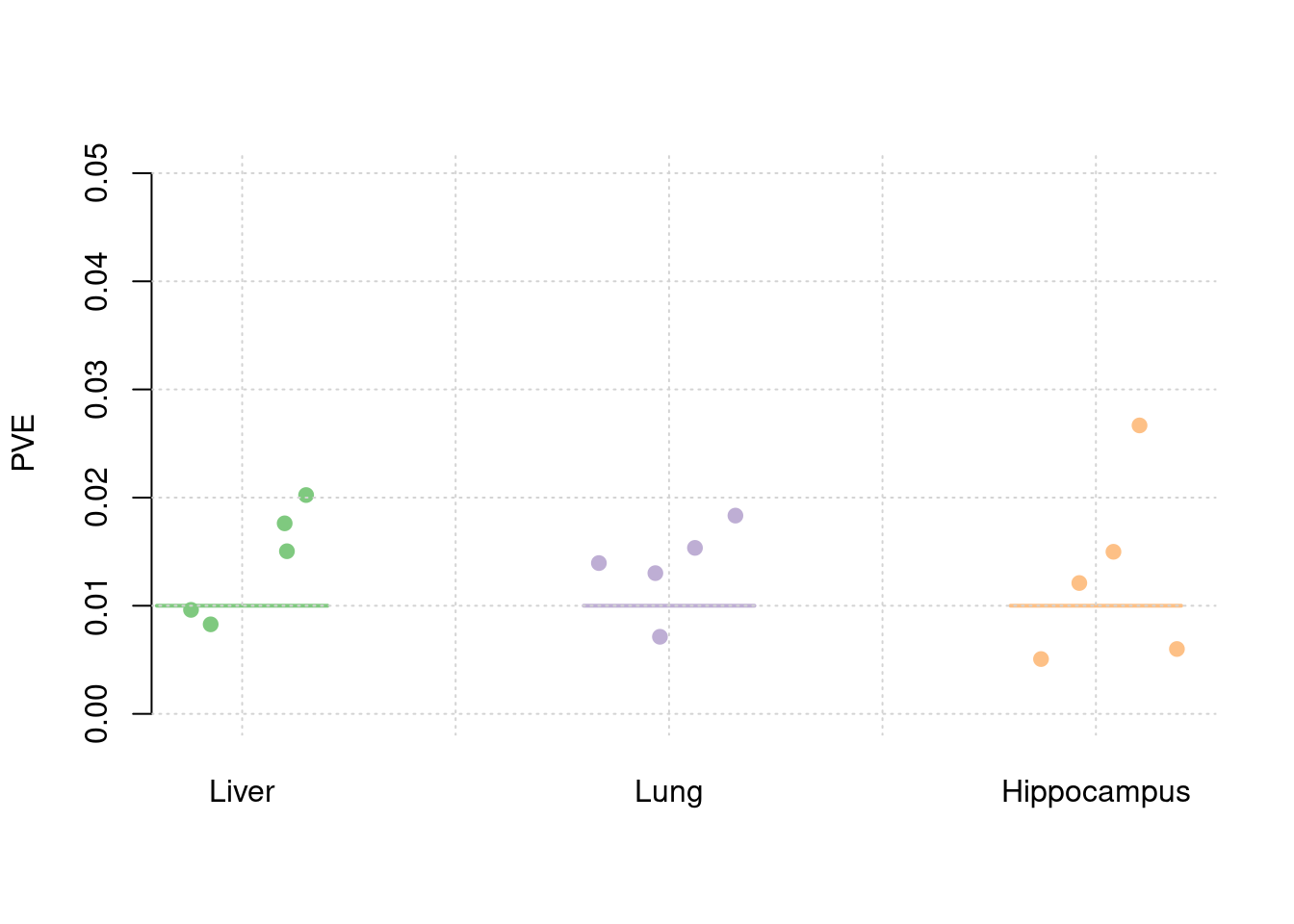
Individual tissue analyses
For the cTWAS analysis, each tissue was analyzed individually and the results were combined.
results_dir <- "/project2/xinhe/shengqian/cTWAS/cTWAS_simulation/simulation_uncorrelated_drop_merge/"
runtag = "ukb-s80.45-3_uncorr"
configtag <- 2
simutags <- paste(2, 1:5, sep = "-")
thin <- 0.1
sample_size <- 45000
PIP_threshold <- 0.8results_df <- get_sim_ind_res(results_dir,runtag,configtag,simutags,thin,sample_size,PIP_threshold)#results using weight1
results_df[,c("simutag", colnames(results_df)[grep("weight1", colnames(results_df))])] simutag n_detected_weight1 n_detected_in_causal_weight1
1 2-1 8 7
2 2-2 11 10
3 2-3 7 7
4 2-4 5 4
5 2-5 5 5#mean percent causal using PIP > 0.8 for weight1
sum(results_df$n_detected_in_causal_weight1)/sum(results_df$n_detected_weight1)[1] 0.9166667#results using weight2
results_df[,c("simutag", colnames(results_df)[grep("weight2", colnames(results_df))])] simutag n_detected_weight2 n_detected_in_causal_weight2
1 2-1 3 2
2 2-2 7 4
3 2-3 7 6
4 2-4 6 5
5 2-5 7 4#mean percent causal using PIP > 0.8 for weight1
sum(results_df$n_detected_in_causal_weight2)/sum(results_df$n_detected_weight2)[1] 0.7#results using weight3
results_df[,c("simutag", colnames(results_df)[grep("weight3", colnames(results_df))])] simutag n_detected_weight3 n_detected_in_causal_weight3
1 2-1 4 4
2 2-2 14 12
3 2-3 8 7
4 2-4 5 4
5 2-5 1 1#mean percent causal using PIP > 0.8 for weight3
sum(results_df$n_detected_in_causal_weight3)/sum(results_df$n_detected_weight3)[1] 0.875#results using combined analysis
results_df[,c("simutag", colnames(results_df)[grep("combined", colnames(results_df))])] simutag n_causal_combined n_detected_combined n_detected_in_causal_combined
1 2-1 66 12 11
2 2-2 80 23 19
3 2-3 60 20 18
4 2-4 70 11 8
5 2-5 67 8 5#mean percent causal using PIP > 0.8 for weight3
sum(results_df$n_detected_in_causal_combined)/sum(results_df$n_detected_combined)[1] 0.8243243Figure

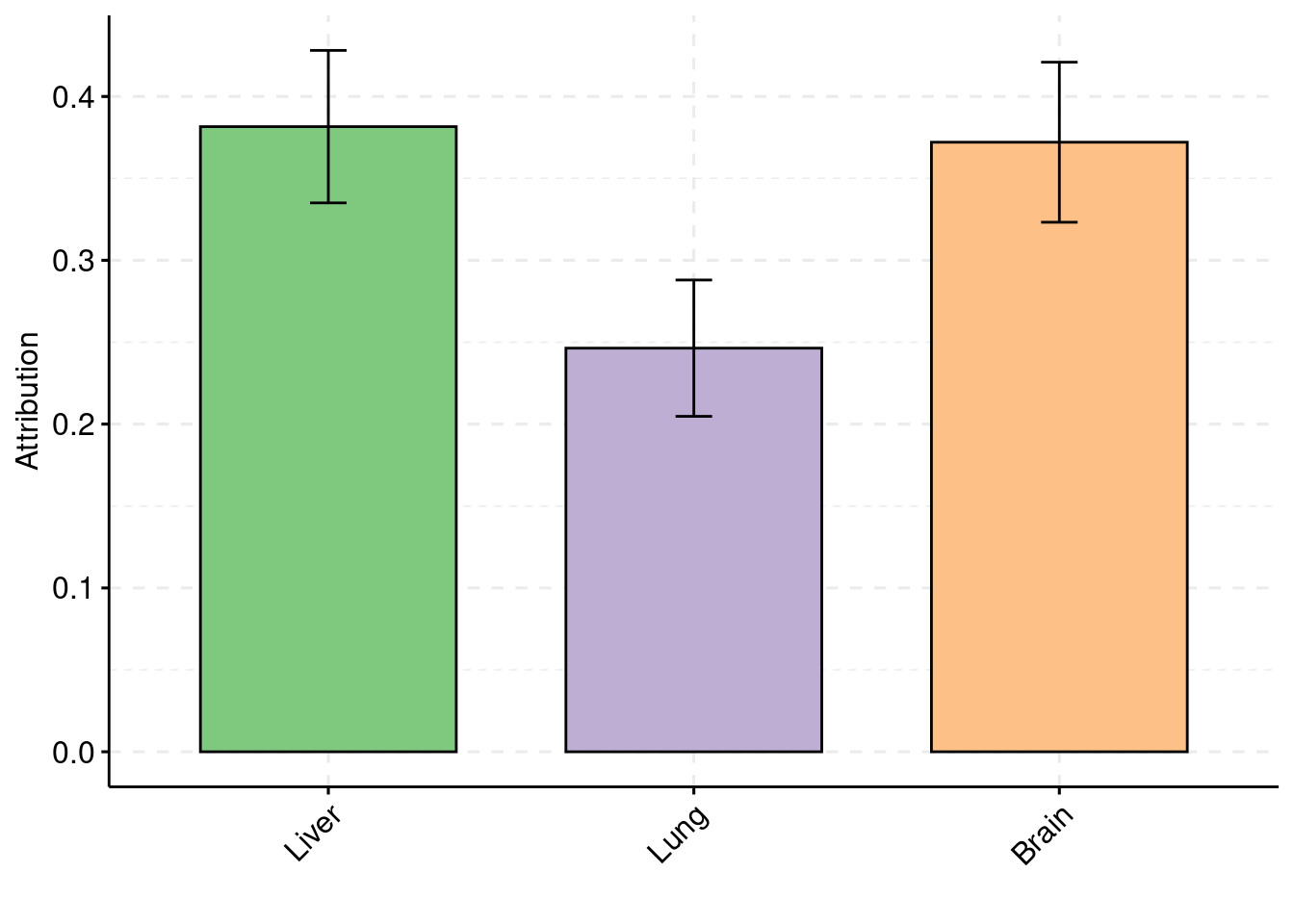
Simulation 3: Three causal tissues with unequal PVE
Separate effect size parameters
30% PVE and 2.5e-4 prior inclusion for SNPs, 3% PVE and 0.009 prior inclusion for Liver expression, 2% PVE and 0.006 prior inclusion for Lung expression, 1% PVE and 0.003 prior inclusion for Brain Hippocampus expression. Each tissue had its own prior inclusion parameter and effect size parameter.
results_dir <- "/project2/xinhe/shengqian/cTWAS/cTWAS_simulation/simulation_uncorrelated_drop_merge/"
runtag = "ukb-s80.45-3_uncorr"
configtag <- 2
simutags <- paste(3, 1:5, sep = "-")
thin <- 0.1
sample_size <- 45000
PIP_threshold <- 0.8results_df <- get_sim_joint_res(results_dir,runtag,configtag,simutags,thin,sample_size,PIP_threshold)#results using PIP threshold (gene+tissue)
results_df[,c("simutag", "n_causal", "n_detected_pip", "n_detected_pip_in_causal")] simutag n_causal n_detected_pip n_detected_pip_in_causal
1 3-1 129 13 12
2 3-2 154 15 13
3 3-3 119 16 15
4 3-4 129 19 18
5 3-5 140 17 15#mean percent causal using PIP > 0.8
sum(results_df$n_detected_pip_in_causal)/sum(results_df$n_detected_pip)[1] 0.9125#results using combined PIP threshold
results_df[,c("simutag", "n_causal_combined", "n_detected_comb_pip", "n_detected_comb_pip_in_causal")] simutag n_causal_combined n_detected_comb_pip n_detected_comb_pip_in_causal
1 3-1 129 26 22
2 3-2 153 30 24
3 3-3 118 22 20
4 3-4 129 35 31
5 3-5 139 27 23#mean percent causal using combined PIP > 0.8
sum(results_df$n_detected_comb_pip_in_causal)/sum(results_df$n_detected_comb_pip)[1] 0.8571429#prior inclusion and mean prior inclusion
results_df[,c(which(colnames(results_df)=="simutag"), setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df))))] simutag prior_snp prior_weight1 prior_weight2 prior_weight3
1 3-1 0.0002595366 0.007402438 0.004632794 0.002491405
2 3-2 0.0002790883 0.005940978 0.004177815 0.011391572
3 3-3 0.0002755165 0.004023845 0.004185074 0.006010862
4 3-4 0.0002173136 0.008609928 0.004372277 0.004850031
5 3-5 0.0002534751 0.010863189 0.008317122 0.003776230colMeans(results_df[,setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df)))]) prior_snp prior_weight1 prior_weight2 prior_weight3
0.000256986 0.007368075 0.005137016 0.005704020 #prior variance and mean prior variance
results_df[,c(which(colnames(results_df)=="simutag"), grep("prior_var", names(results_df)))] simutag prior_var_snp prior_var_weight1 prior_var_weight2 prior_var_weight3
1 3-1 8.218313 15.99248 10.340997 18.268167
2 3-2 7.111651 26.20390 13.757128 12.163354
3 3-3 7.785969 40.83585 13.481862 13.289472
4 3-4 9.186761 23.66419 21.154161 9.848333
5 3-5 7.832367 23.24540 5.908057 17.928854colMeans(results_df[,grep("prior_var", names(results_df))]) prior_var_snp prior_var_weight1 prior_var_weight2 prior_var_weight3
8.027012 25.988364 12.928441 14.299636 #PVE and mean PVE
results_df[,c(which(colnames(results_df)=="simutag"), grep("pve", names(results_df)))] simutag pve_snp pve_weight1 pve_weight2 pve_weight3
1 3-1 0.2510808 0.01945432 0.01040555 0.008051828
2 3-2 0.2336384 0.02558289 0.01248351 0.024512754
3 3-3 0.2525181 0.02700278 0.01225499 0.014131868
4 3-4 0.2350074 0.03348242 0.02008923 0.008450109
5 3-5 0.2337010 0.04149733 0.01067278 0.011977498colMeans(results_df[,grep("pve", names(results_df))]) pve_snp pve_weight1 pve_weight2 pve_weight3
0.24118915 0.02940395 0.01318121 0.01342481 #TWAS results
results_df[,c(which(colnames(results_df)=="simutag"), grep("twas", names(results_df)))] simutag n_detected_twas n_detected_twas_in_causal n_detected_comb_twas
1 3-1 260 34 159
2 3-2 217 38 143
3 3-3 200 33 134
4 3-4 197 36 111
5 3-5 243 39 148
n_detected_comb_twas_in_causal
1 36
2 38
3 34
4 36
5 39sum(results_df$n_detected_comb_twas_in_causal)/sum(results_df$n_detected_comb_twas)[1] 0.2633094y1 <- results_df$prior_weight1
y2 <- results_df$prior_weight2
y3 <- results_df$prior_weight3
truth <- rbind(c(1,0.009),c(2,0.006),c(3,0.003))
est <- rbind(cbind(1,y1),cbind(2,y2),cbind(3,y3))
plot_par(truth,est,xlabels = c("Liver","Lung","Hippocampus"),ylim=c(0,0.025),ylab="Prior inclusion")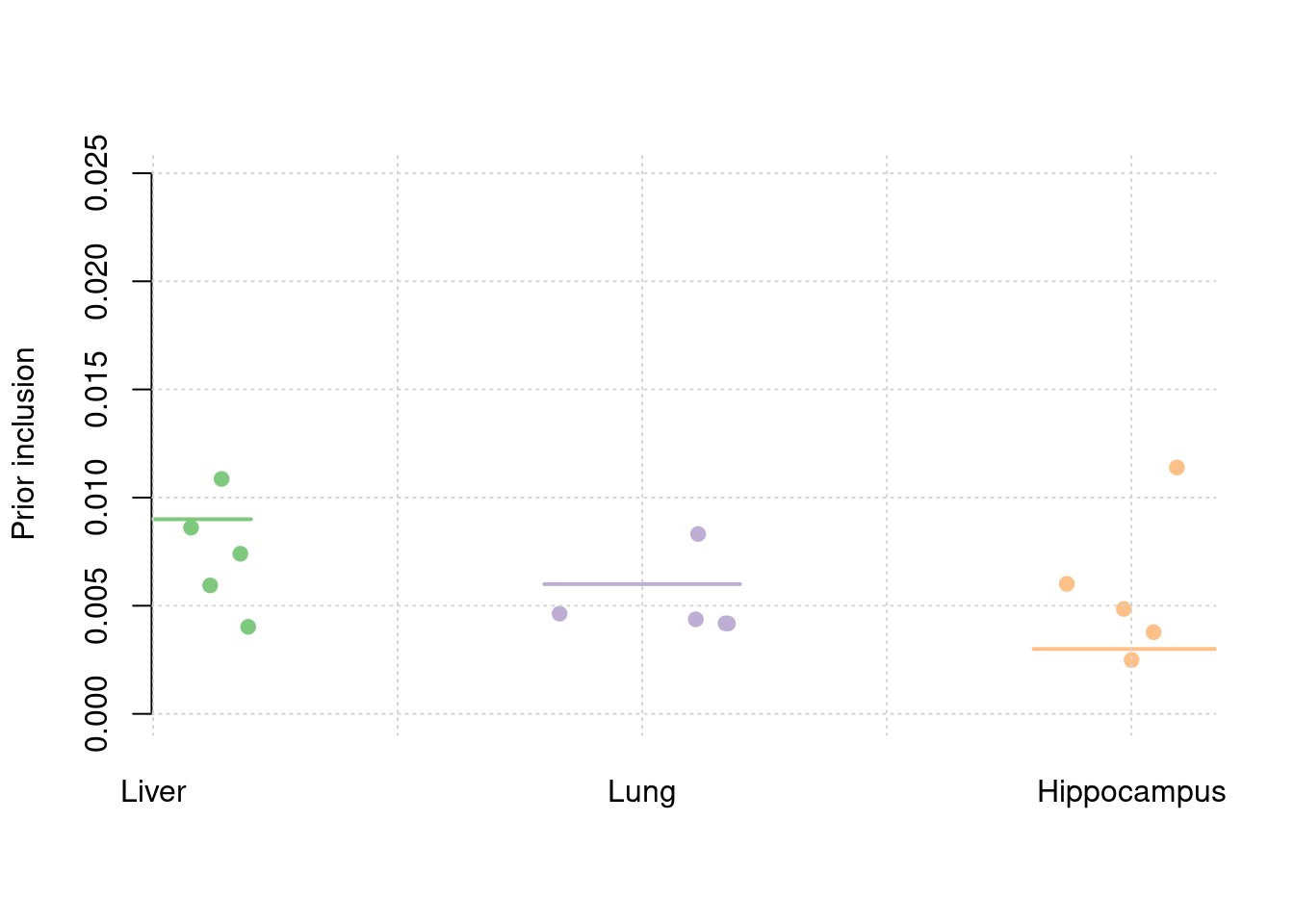
| Version | Author | Date |
|---|---|---|
| f296597 | sq-96 | 2023-10-20 |
y1 <- results_df$prior_weight1/results_df$prior_snp
y2 <- results_df$prior_weight2/results_df$prior_snp
y3 <- results_df$prior_weight3/results_df$prior_snp
truth <- rbind(c(1,36),c(2,24),c(3,12))
est <- rbind(cbind(1,y1),cbind(2,y2),cbind(3,y3))
plot_par(truth,est,xlabels = c("Liver","Lung","Hippocampus"),ylim=c(0,60),ylab="Enrichment")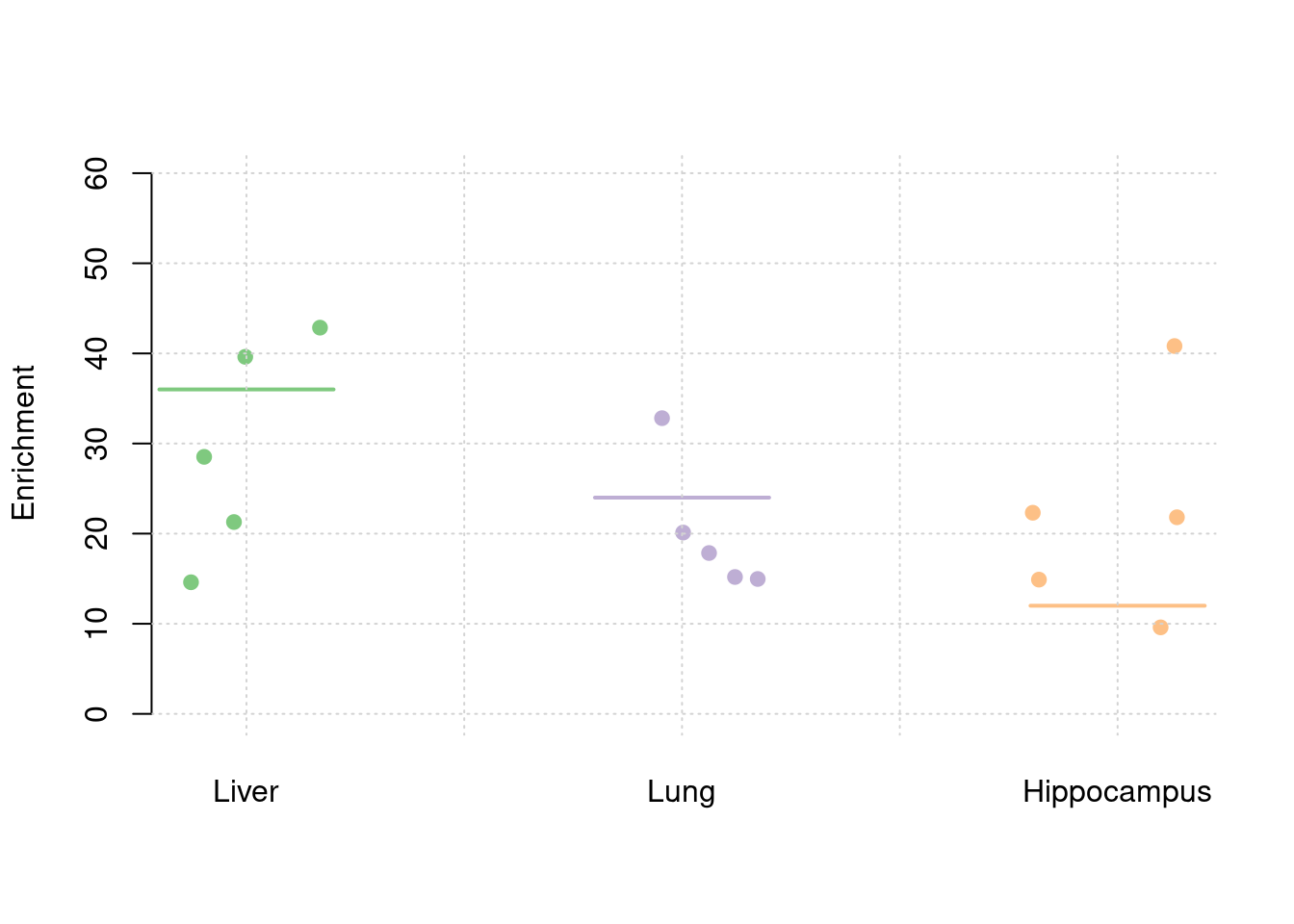
y1 <- results_df$pve_weight1
y2 <- results_df$pve_weight2
y3 <- results_df$pve_weight3
truth <- rbind(c(1,0.03),c(2,0.02),c(3,0.01))
est <- rbind(cbind(1,y1),cbind(2,y2),cbind(3,y3))
plot_par(truth,est,xlabels = c("Liver","Lung","Hippocampus"),ylim=c(0,0.05),ylab="PVE")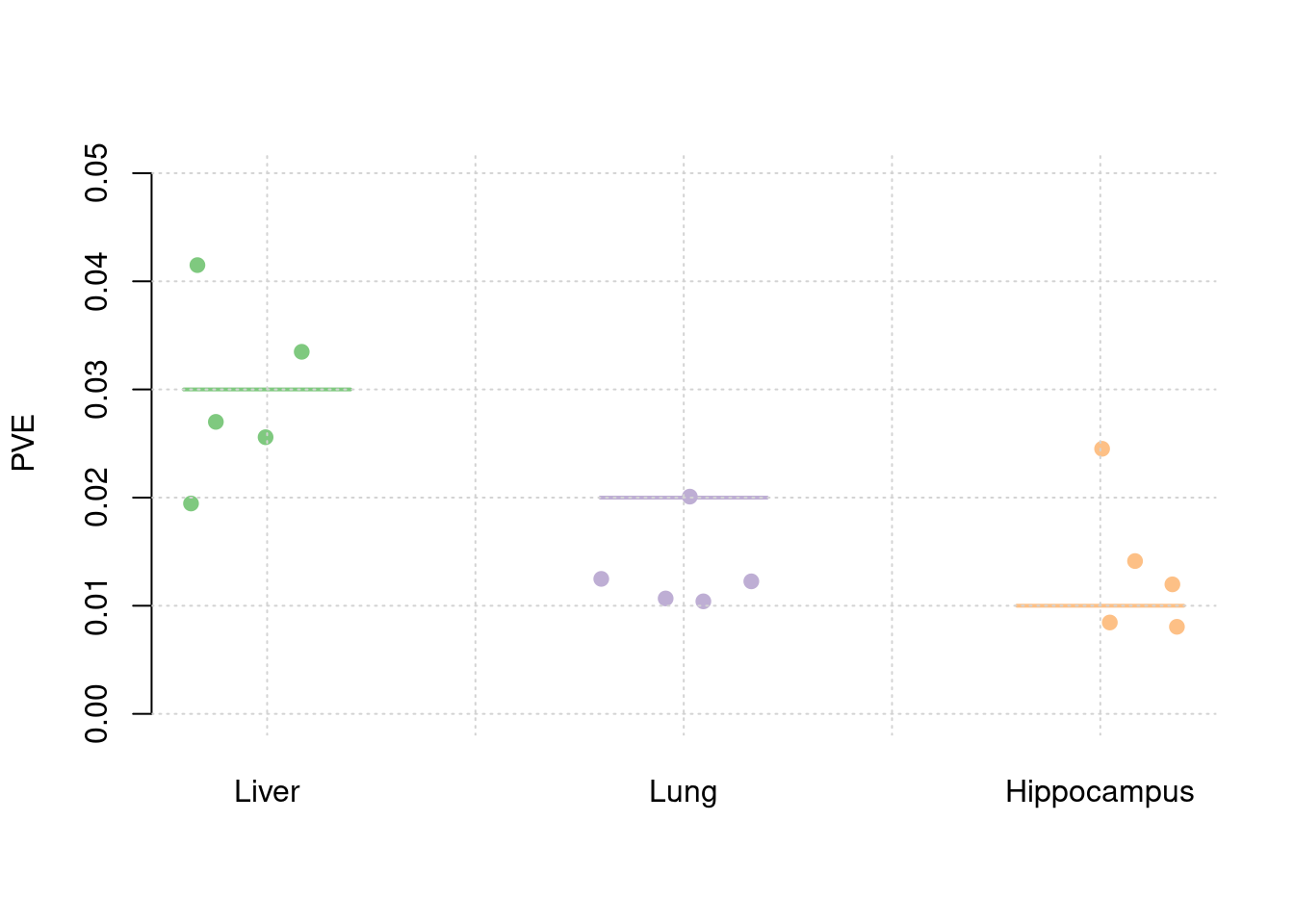
Individual tissue analyses
For the cTWAS analysis, each tissue was analyzed individually and the results were combined.
results_dir <- "/project2/xinhe/shengqian/cTWAS/cTWAS_simulation/simulation_uncorrelated_drop_merge/"
runtag = "ukb-s80.45-3_uncorr"
configtag <- 2
simutags <- paste(3, 1:5, sep = "-")
thin <- 0.1
sample_size <- 45000
PIP_threshold <- 0.8results_df <- get_sim_ind_res(results_dir,runtag,configtag,simutags,thin,sample_size,PIP_threshold)#results using weight1
results_df[,c("simutag", colnames(results_df)[grep("weight1", colnames(results_df))])] simutag n_detected_weight1 n_detected_in_causal_weight1
1 3-1 20 16
2 3-2 20 16
3 3-3 12 12
4 3-4 23 20
5 3-5 18 16#mean percent causal using PIP > 0.8 for weight1
sum(results_df$n_detected_in_causal_weight1)/sum(results_df$n_detected_weight1)[1] 0.8602151#results using weight2
results_df[,c("simutag", colnames(results_df)[grep("weight2", colnames(results_df))])] simutag n_detected_weight2 n_detected_in_causal_weight2
1 3-1 9 7
2 3-2 14 10
3 3-3 7 7
4 3-4 14 13
5 3-5 9 9#mean percent causal using PIP > 0.8 for weight1
sum(results_df$n_detected_in_causal_weight2)/sum(results_df$n_detected_weight2)[1] 0.8679245#results using weight3
results_df[,c("simutag", colnames(results_df)[grep("weight3", colnames(results_df))])] simutag n_detected_weight3 n_detected_in_causal_weight3
1 3-1 7 7
2 3-2 15 11
3 3-3 4 3
4 3-4 13 11
5 3-5 6 5#mean percent causal using PIP > 0.8 for weight3
sum(results_df$n_detected_in_causal_weight3)/sum(results_df$n_detected_weight3)[1] 0.8222222#results using combined analysis
results_df[,c("simutag", colnames(results_df)[grep("combined", colnames(results_df))])] simutag n_causal_combined n_detected_combined n_detected_in_causal_combined
1 3-1 129 29 24
2 3-2 153 32 24
3 3-3 118 20 19
4 3-4 129 37 31
5 3-5 139 26 24#mean percent causal using PIP > 0.8 for weight3
sum(results_df$n_detected_in_causal_combined)/sum(results_df$n_detected_combined)[1] 0.8472222Figure
For the cTWAS analysis, each tissue was analyzed individually and the results were combined. 
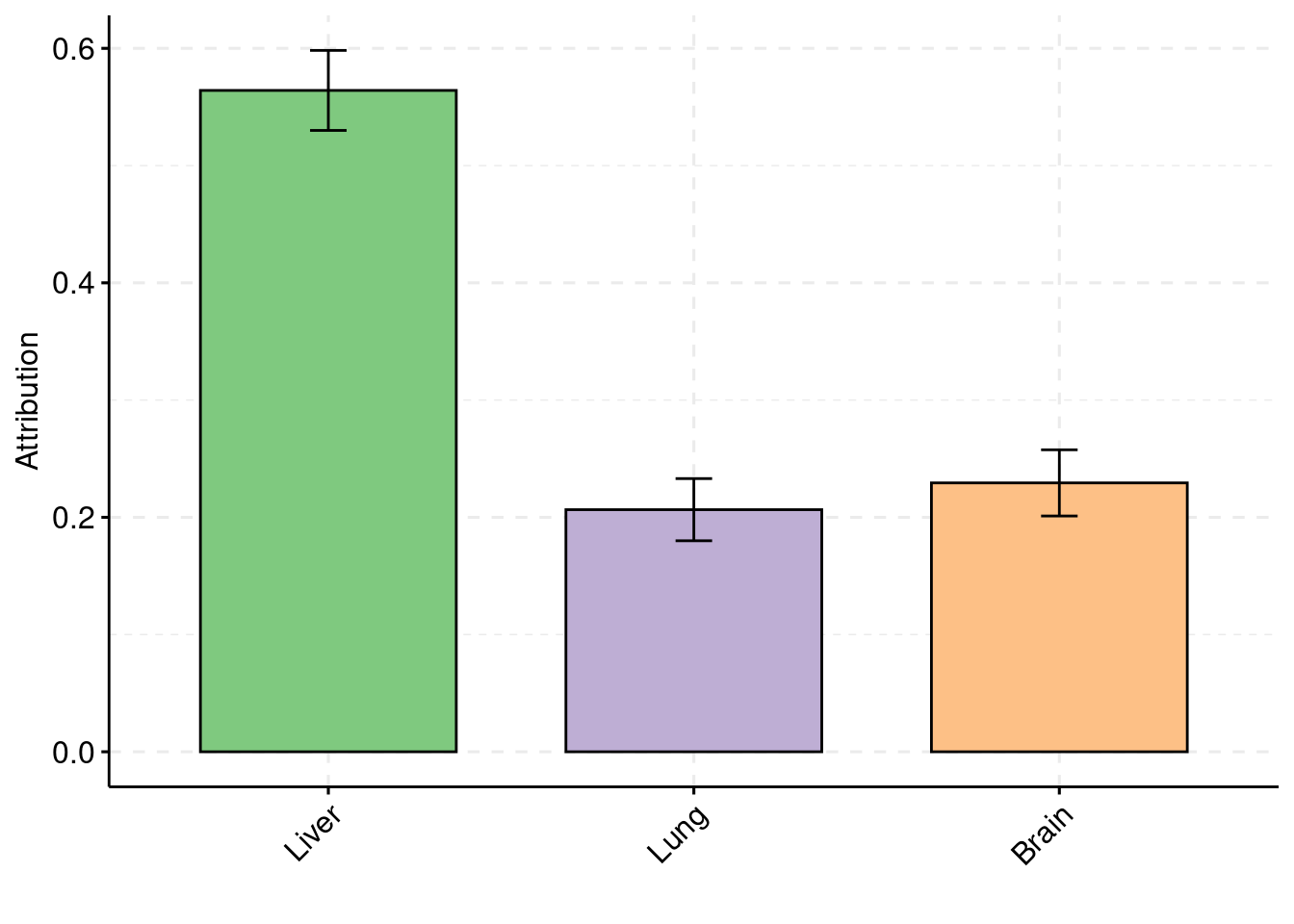 ## Simulation 4: Two causal tissues with unequal PVE and one non-causal tissue
## Simulation 4: Two causal tissues with unequal PVE and one non-causal tissue
Separate effect size parameters
30% PVE and 2.5e-4 prior inclusion for SNPs, 3% PVE and 0.009 prior inclusion for Liver expression, 1% PVE and 0.003 prior inclusion for Lung expression, Brain Hippocampus expression has no effects. Each tissue had its own prior inclusion parameter and effect size parameter.
results_dir <- "/project2/xinhe/shengqian/cTWAS/cTWAS_simulation/simulation_uncorrelated_drop_merge/"
runtag = "ukb-s80.45-3_uncorr"
configtag <- 2
simutags <- paste(4, 1:5, sep = "-")
thin <- 0.1
sample_size <- 45000
PIP_threshold <- 0.8results_df <- get_sim_joint_res(results_dir,runtag,configtag,simutags,thin,sample_size,PIP_threshold)#results using PIP threshold (gene+tissue)
results_df[,c("simutag", "n_causal", "n_detected_pip", "n_detected_pip_in_causal")] simutag n_causal n_detected_pip n_detected_pip_in_causal
1 4-1 85 14 14
2 4-2 93 15 14
3 4-3 116 22 21
4 4-4 82 13 13
5 4-5 99 11 10#mean percent causal using PIP > 0.8
sum(results_df$n_detected_pip_in_causal)/sum(results_df$n_detected_pip)[1] 0.96#results using combined PIP threshold
results_df[,c("simutag", "n_causal_combined", "n_detected_comb_pip", "n_detected_comb_pip_in_causal")] simutag n_causal_combined n_detected_comb_pip n_detected_comb_pip_in_causal
1 4-1 85 19 18
2 4-2 93 23 18
3 4-3 116 33 30
4 4-4 82 14 14
5 4-5 99 16 13#mean percent causal using combined PIP > 0.8
sum(results_df$n_detected_comb_pip_in_causal)/sum(results_df$n_detected_comb_pip)[1] 0.8857143#prior inclusion and mean prior inclusion
results_df[,c(which(colnames(results_df)=="simutag"), setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df))))] simutag prior_snp prior_weight1 prior_weight2 prior_weight3
1 4-1 0.0002836507 0.007823094 0.002552276 0.0031298371
2 4-2 0.0002505663 0.004055552 0.003984521 0.0025329456
3 4-3 0.0002791923 0.012775958 0.004421434 0.0023033326
4 4-4 0.0002728597 0.008072991 0.003259917 0.0022185325
5 4-5 0.0002469015 0.007428323 0.007842161 0.0009239835colMeans(results_df[,setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df)))]) prior_snp prior_weight1 prior_weight2 prior_weight3
0.0002666341 0.0080311834 0.0044120619 0.0022217263 #prior variance and mean prior variance
results_df[,c(which(colnames(results_df)=="simutag"), grep("prior_var", names(results_df)))] simutag prior_var_snp prior_var_weight1 prior_var_weight2 prior_var_weight3
1 4-1 7.316408 31.15189 17.048708 7.159584
2 4-2 8.034003 32.73799 16.948387 41.189388
3 4-3 7.762142 16.82626 19.447154 2.698047
4 4-4 8.071062 24.48370 6.994372 3.304820
5 4-5 8.015189 29.22436 4.708917 1.167933colMeans(results_df[,grep("prior_var", names(results_df))]) prior_var_snp prior_var_weight1 prior_var_weight2 prior_var_weight3
7.839761 26.884840 13.029507 11.103954 #PVE and mean PVE
results_df[,c(which(colnames(results_df)=="simutag"), grep("pve", names(results_df)))] simutag pve_snp pve_weight1 pve_weight2 pve_weight3
1 4-1 0.2442947 0.04004872 0.009451025 0.0039642830
2 4-2 0.2369665 0.02181864 0.014667778 0.0184572210
3 4-3 0.2551040 0.03532700 0.018675793 0.0010994138
4 4-4 0.2592402 0.03248158 0.004952393 0.0012970858
5 4-5 0.2329537 0.03567480 0.008020779 0.0001909137colMeans(results_df[,grep("pve", names(results_df))]) pve_snp pve_weight1 pve_weight2 pve_weight3
0.245711822 0.033070145 0.011153554 0.005001783 #TWAS results
results_df[,c(which(colnames(results_df)=="simutag"), grep("twas", names(results_df)))] simutag n_detected_twas n_detected_twas_in_causal n_detected_comb_twas
1 4-1 153 24 98
2 4-2 214 30 134
3 4-3 138 32 92
4 4-4 111 21 76
5 4-5 169 22 106
n_detected_comb_twas_in_causal
1 24
2 30
3 32
4 21
5 22sum(results_df$n_detected_comb_twas_in_causal)/sum(results_df$n_detected_comb_twas)[1] 0.2549407y1 <- results_df$prior_weight1
y2 <- results_df$prior_weight2
y3 <- results_df$prior_weight3
truth <- rbind(c(1,0.009),c(2,0.003),c(3,0))
est <- rbind(cbind(1,y1),cbind(2,y2),cbind(3,y3))
plot_par(truth,est,xlabels = c("Liver","Lung","Hippocampus"),ylim=c(0,0.025),ylab="Prior inclusion")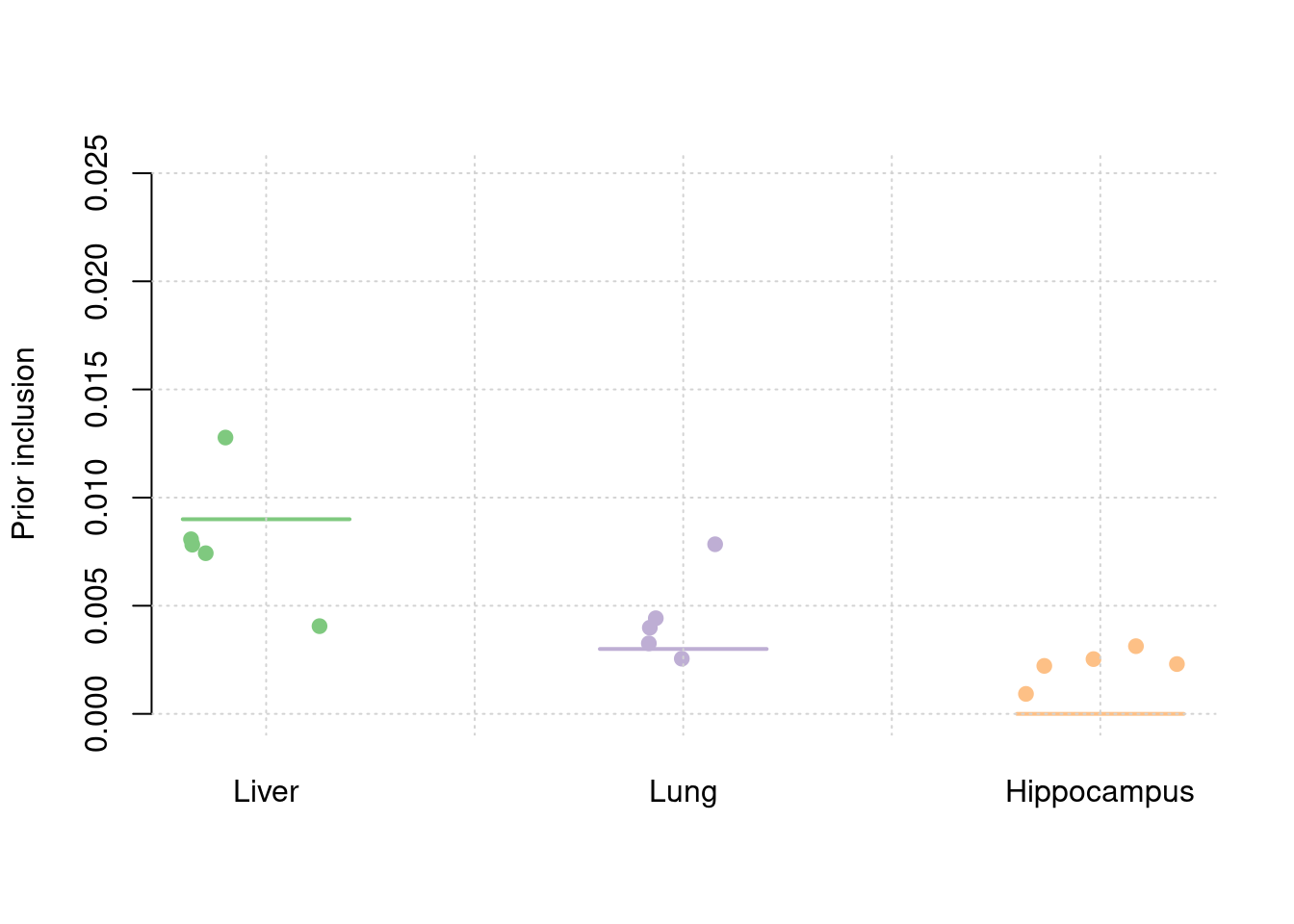
y1 <- results_df$prior_weight1/results_df$prior_snp
y2 <- results_df$prior_weight2/results_df$prior_snp
y3 <- results_df$prior_weight3/results_df$prior_snp
truth <- rbind(c(1,36),c(2,12),c(3,0))
est <- rbind(cbind(1,y1),cbind(2,y2),cbind(3,y3))
plot_par(truth,est,xlabels = c("Liver","Lung","Hippocampus"),ylim=c(0,60),ylab="Enrichment")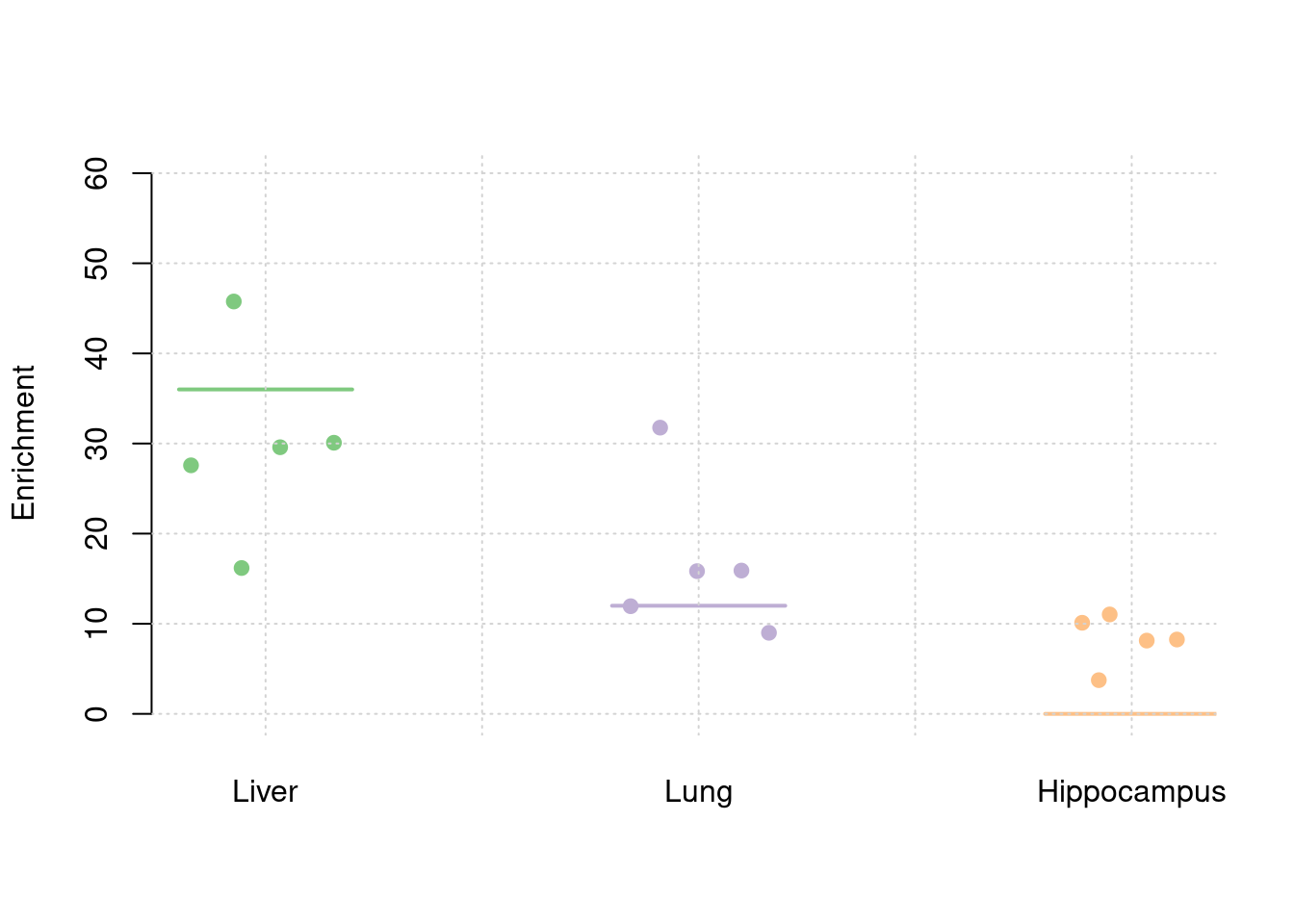
y1 <- results_df$pve_weight1
y2 <- results_df$pve_weight2
y3 <- results_df$pve_weight3
truth <- rbind(c(1,0.03),c(2,0.01),c(3,0))
est <- rbind(cbind(1,y1),cbind(2,y2),cbind(3,y3))
plot_par(truth,est,xlabels = c("Liver","Lung","Hippocampus"),ylim=c(0,0.05),ylab="PVE")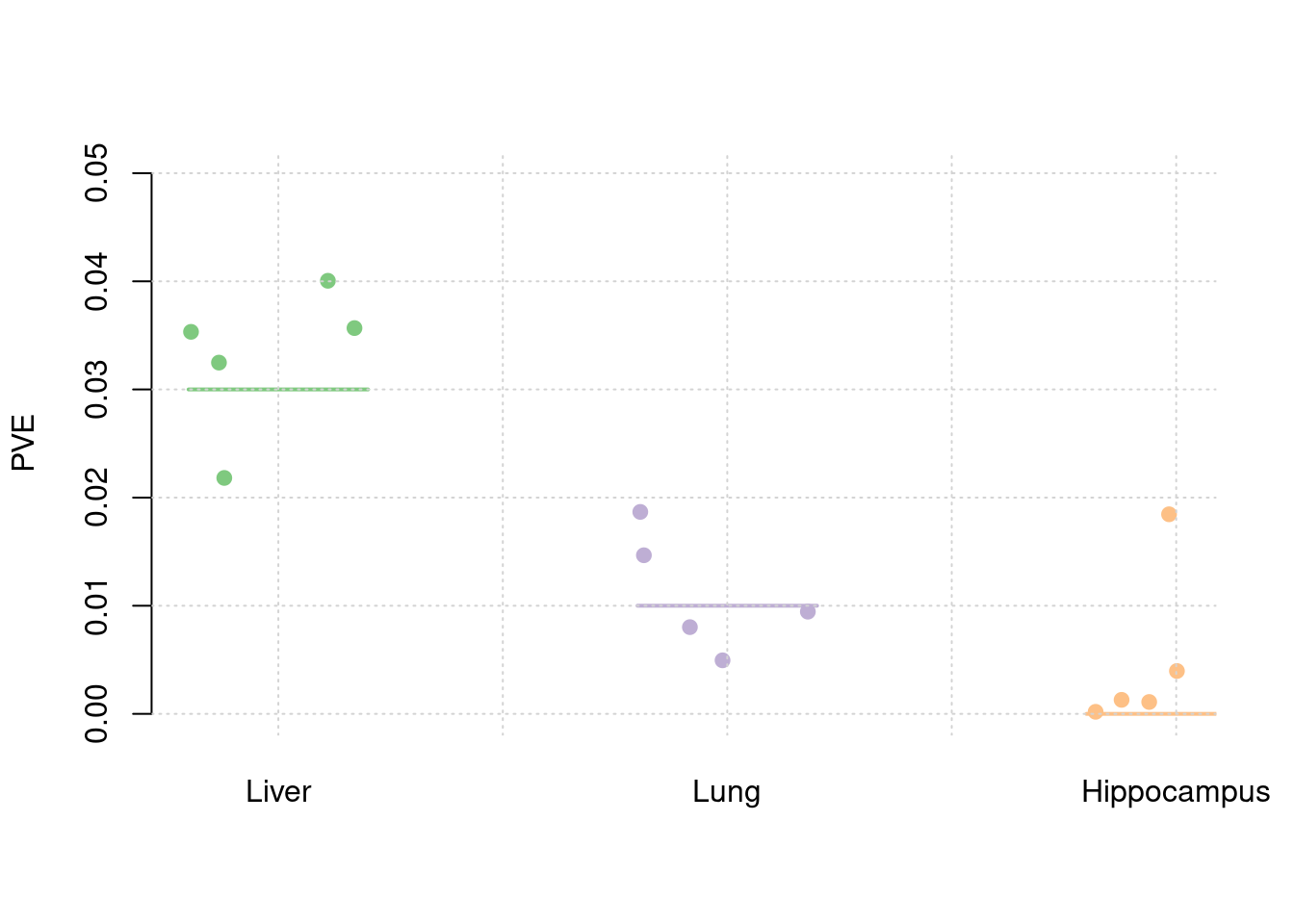
Individual tissue analyses
For the cTWAS analysis, each tissue was analyzed individually and the results were combined.
results_dir <- "/project2/xinhe/shengqian/cTWAS/cTWAS_simulation/simulation_uncorrelated_drop_merge/"
runtag = "ukb-s80.45-3_uncorr"
configtag <- 2
simutags <- paste(4, 1:5, sep = "-")
thin <- 0.1
sample_size <- 45000
PIP_threshold <- 0.8results_df <- get_sim_ind_res(results_dir,runtag,configtag,simutags,thin,sample_size,PIP_threshold)#results using weight1
results_df[,c("simutag", colnames(results_df)[grep("weight1", colnames(results_df))])] simutag n_detected_weight1 n_detected_in_causal_weight1
1 4-1 14 14
2 4-2 21 18
3 4-3 24 21
4 4-4 13 13
5 4-5 17 14#mean percent causal using PIP > 0.8 for weight1
sum(results_df$n_detected_in_causal_weight1)/sum(results_df$n_detected_weight1)[1] 0.8988764#results using weight2
results_df[,c("simutag", colnames(results_df)[grep("weight2", colnames(results_df))])] simutag n_detected_weight2 n_detected_in_causal_weight2
1 4-1 3 3
2 4-2 9 8
3 4-3 14 14
4 4-4 4 4
5 4-5 5 1#mean percent causal using PIP > 0.8 for weight1
sum(results_df$n_detected_in_causal_weight2)/sum(results_df$n_detected_weight2)[1] 0.8571429#results using weight3
results_df[,c("simutag", colnames(results_df)[grep("weight3", colnames(results_df))])] simutag n_detected_weight3 n_detected_in_causal_weight3
1 4-1 4 3
2 4-2 9 5
3 4-3 3 2
4 4-4 4 4
5 4-5 2 1#mean percent causal using PIP > 0.8 for weight3
sum(results_df$n_detected_in_causal_weight3)/sum(results_df$n_detected_weight3)[1] 0.6818182#results using combined analysis
results_df[,c("simutag", colnames(results_df)[grep("combined", colnames(results_df))])] simutag n_causal_combined n_detected_combined n_detected_in_causal_combined
1 4-1 85 17 16
2 4-2 93 27 21
3 4-3 116 33 29
4 4-4 82 16 16
5 4-5 99 21 15#mean percent causal using PIP > 0.8 for weight3
sum(results_df$n_detected_in_causal_combined)/sum(results_df$n_detected_combined)[1] 0.8508772Figure
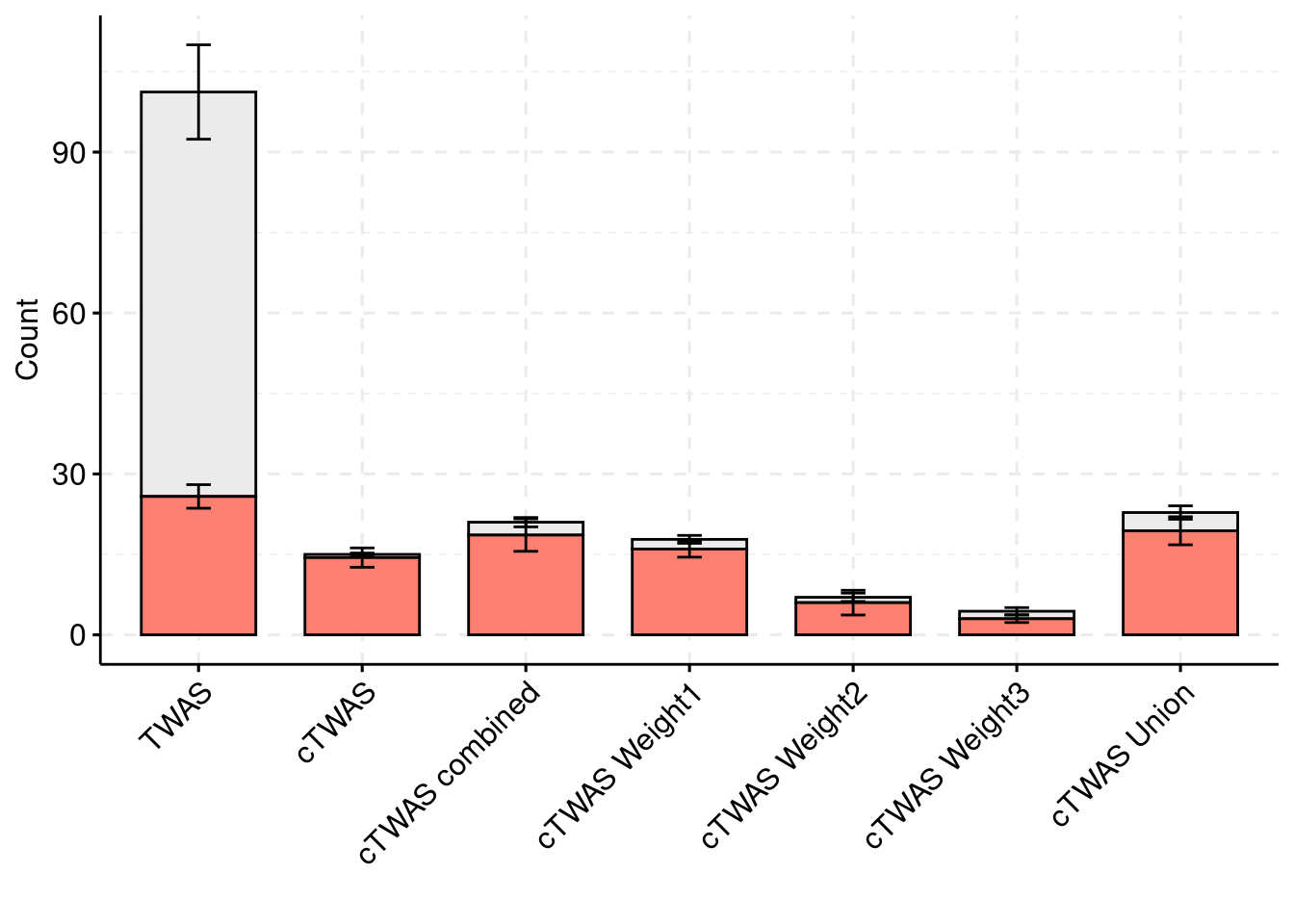
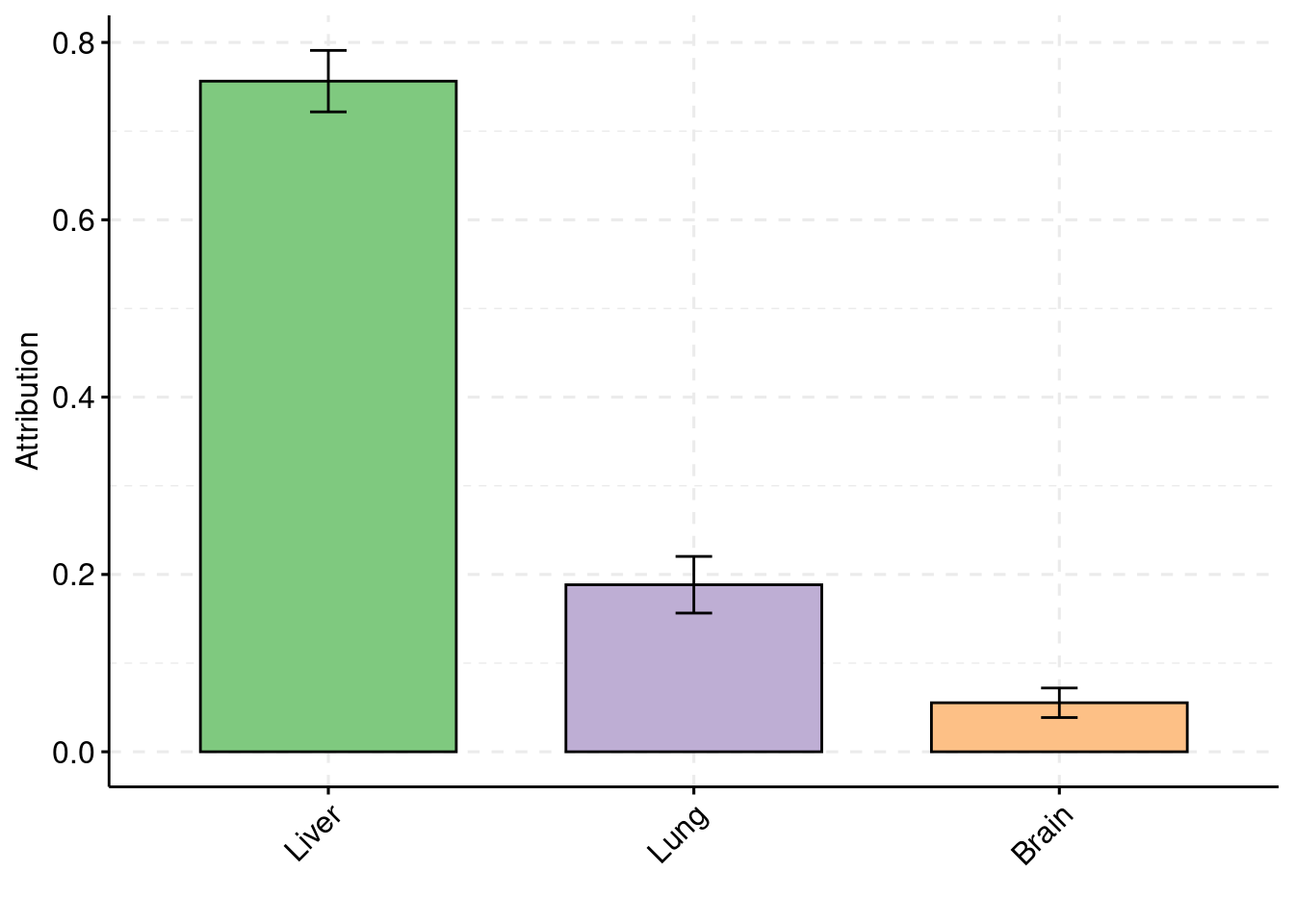
Simulation 5: one causal tissues and two non-causal tissues
Separate effect size parameters
30% PVE and 2.5e-4 prior inclusion for SNPs, 3% PVE and 0.009 prior inclusion for Liver expression, Lung expression and Brain Hippocampus expression has no effects. Each tissue had its own prior inclusion parameter and effect size parameter.
results_dir <- "/project2/xinhe/shengqian/cTWAS/cTWAS_simulation/simulation_uncorrelated_drop_merge/"
runtag = "ukb-s80.45-3_uncorr"
configtag <- 2
simutags <- paste(5, 1:5, sep = "-")
thin <- 0.1
sample_size <- 45000
PIP_threshold <- 0.8results_df <- get_sim_joint_res(results_dir,runtag,configtag,simutags,thin,sample_size,PIP_threshold)#results using PIP threshold (gene+tissue)
results_df[,c("simutag", "n_causal", "n_detected_pip", "n_detected_pip_in_causal")] simutag n_causal n_detected_pip n_detected_pip_in_causal
1 5-1 68 12 12
2 5-2 63 11 10
3 5-3 78 20 17
4 5-4 66 10 8
5 5-5 54 8 6#mean percent causal using PIP > 0.8
sum(results_df$n_detected_pip_in_causal)/sum(results_df$n_detected_pip)[1] 0.8688525#results using combined PIP threshold
results_df[,c("simutag", "n_causal_combined", "n_detected_comb_pip", "n_detected_comb_pip_in_causal")] simutag n_causal_combined n_detected_comb_pip n_detected_comb_pip_in_causal
1 5-1 68 19 19
2 5-2 63 15 13
3 5-3 78 24 21
4 5-4 66 20 15
5 5-5 54 11 8#mean percent causal using combined PIP > 0.8
sum(results_df$n_detected_comb_pip_in_causal)/sum(results_df$n_detected_comb_pip)[1] 0.8539326#prior inclusion and mean prior inclusion
results_df[,c(which(colnames(results_df)=="simutag"), setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df))))] simutag prior_snp prior_weight1 prior_weight2 prior_weight3
1 5-1 0.0002260741 0.012487622 0.006394432 0.002248323
2 5-2 0.0002520776 0.008855700 0.001506860 0.001028561
3 5-3 0.0002604092 0.015701737 0.002744097 0.001945911
4 5-4 0.0002723394 0.008886928 0.005554238 0.004179820
5 5-5 0.0002953418 0.004996529 0.002761793 0.001641676colMeans(results_df[,setdiff(grep("prior", names(results_df)), grep("prior_var", names(results_df)))]) prior_snp prior_weight1 prior_weight2 prior_weight3
0.0002612484 0.0101857033 0.0037922838 0.0022088580 #prior variance and mean prior variance
results_df[,c(which(colnames(results_df)=="simutag"), grep("prior_var", names(results_df)))] simutag prior_var_snp prior_var_weight1 prior_var_weight2 prior_var_weight3
1 5-1 9.701863 12.98372 5.581177 17.569708
2 5-2 9.089926 22.74965 8.864505 10.116937
3 5-3 8.942667 16.42152 8.574377 3.018352
4 5-4 8.244432 15.40885 6.278919 9.578499
5 5-5 7.343877 20.37894 11.734378 2.023874colMeans(results_df[,grep("prior_var", names(results_df))]) prior_var_snp prior_var_weight1 prior_var_weight2 prior_var_weight3
8.664553 17.588533 8.206671 8.461474 #PVE and mean PVE
results_df[,c(which(colnames(results_df)=="simutag"), grep("pve", names(results_df)))] simutag pve_snp pve_weight1 pve_weight2 pve_weight3
1 5-1 0.2581893 0.02664432 0.007751532 0.0069884089
2 5-2 0.2697285 0.03310726 0.002901263 0.0018409162
3 5-3 0.2741294 0.04237274 0.005110482 0.0010390776
4 5-4 0.2643038 0.02250336 0.007574765 0.0070828842
5 5-5 0.2553187 0.01673307 0.007039000 0.0005877952colMeans(results_df[,grep("pve", names(results_df))]) pve_snp pve_weight1 pve_weight2 pve_weight3
0.264333936 0.028272151 0.006075409 0.003507816 #TWAS results
results_df[,c(which(colnames(results_df)=="simutag"), grep("twas", names(results_df)))] simutag n_detected_twas n_detected_twas_in_causal n_detected_comb_twas
1 5-1 111 21 77
2 5-2 145 19 84
3 5-3 190 33 118
4 5-4 145 19 93
5 5-5 98 15 65
n_detected_comb_twas_in_causal
1 21
2 19
3 33
4 20
5 16sum(results_df$n_detected_comb_twas_in_causal)/sum(results_df$n_detected_comb_twas)[1] 0.2494279y1 <- results_df$prior_weight1
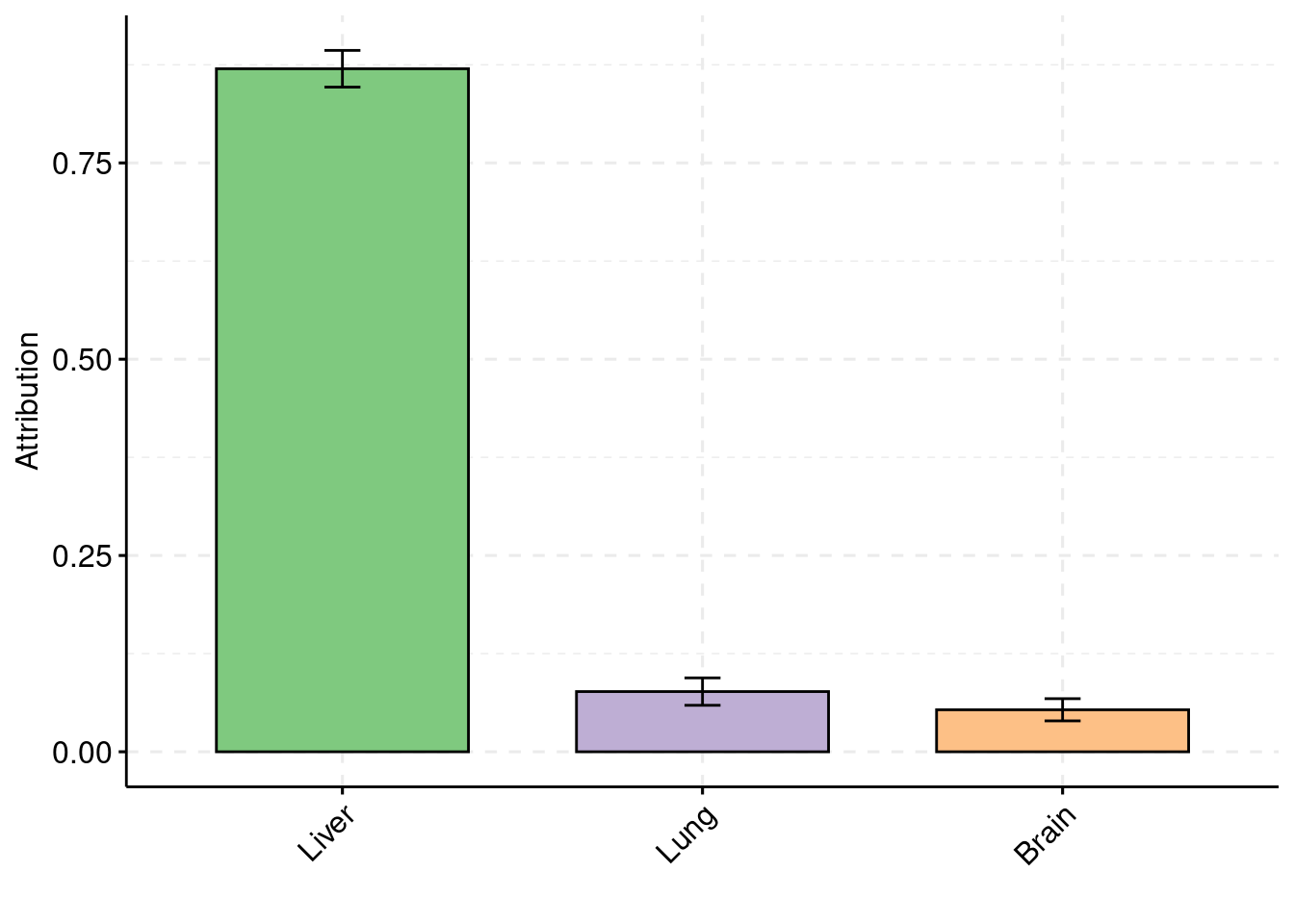
y2 <- results_df$prior_weight2
y3 <- results_df$prior_weight3
truth <- rbind(c(1,0.009),c(2,0),c(3,0))
est <- rbind(cbind(1,y1),cbind(2,y2),cbind(3,y3))
plot_par(truth,est,xlabels = c("Liver","Lung","Hippocampus"),ylim=c(0,0.025),ylab="Prior inclusion")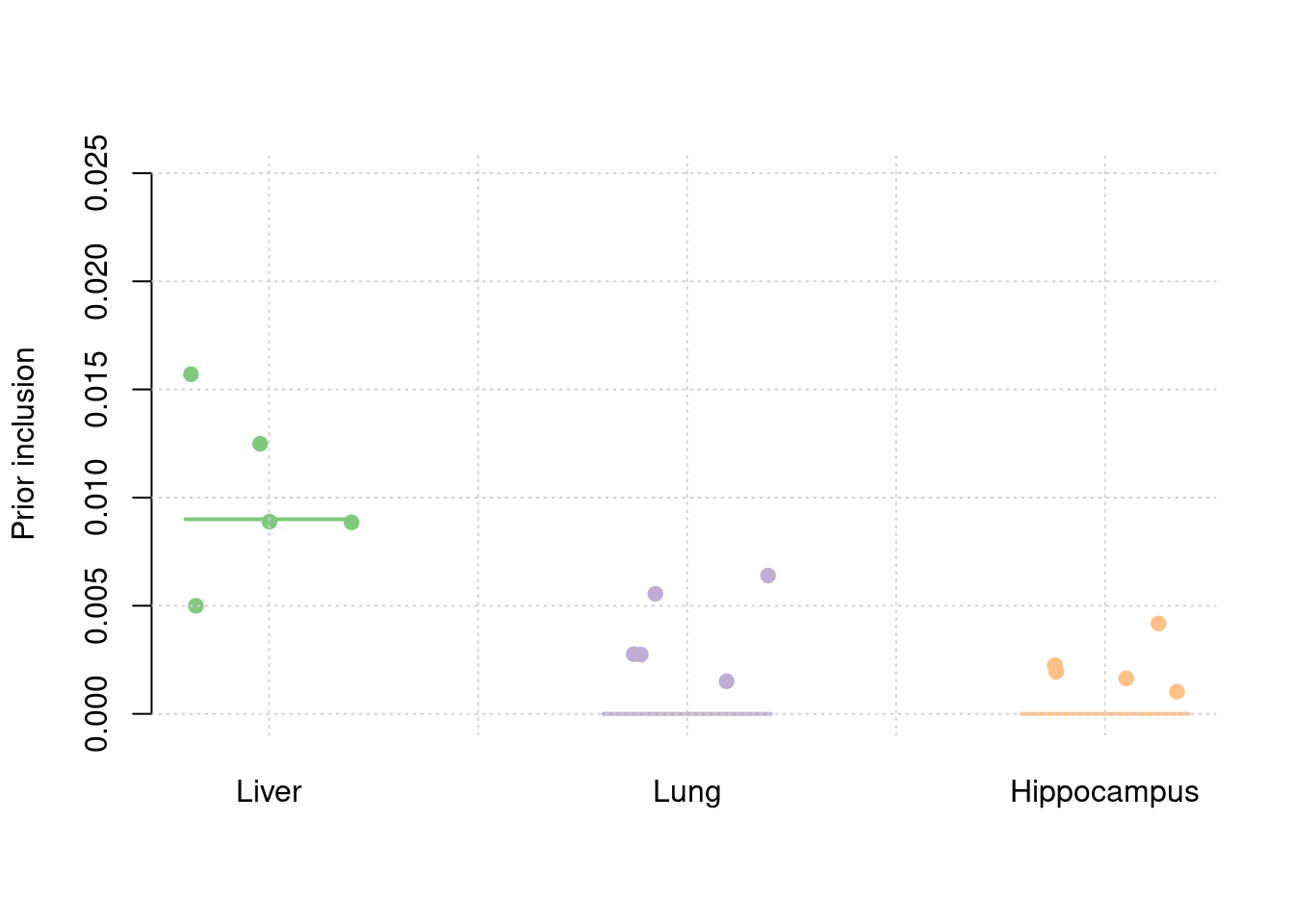
y1 <- results_df$prior_weight1/results_df$prior_snp
y2 <- results_df$prior_weight2/results_df$prior_snp
y3 <- results_df$prior_weight3/results_df$prior_snp
truth <- rbind(c(1,36),c(2,0),c(3,0))
est <- rbind(cbind(1,y1),cbind(2,y2),cbind(3,y3))
plot_par(truth,est,xlabels = c("Liver","Lung","Hippocampus"),ylim=c(0,60),ylab="Enrichment")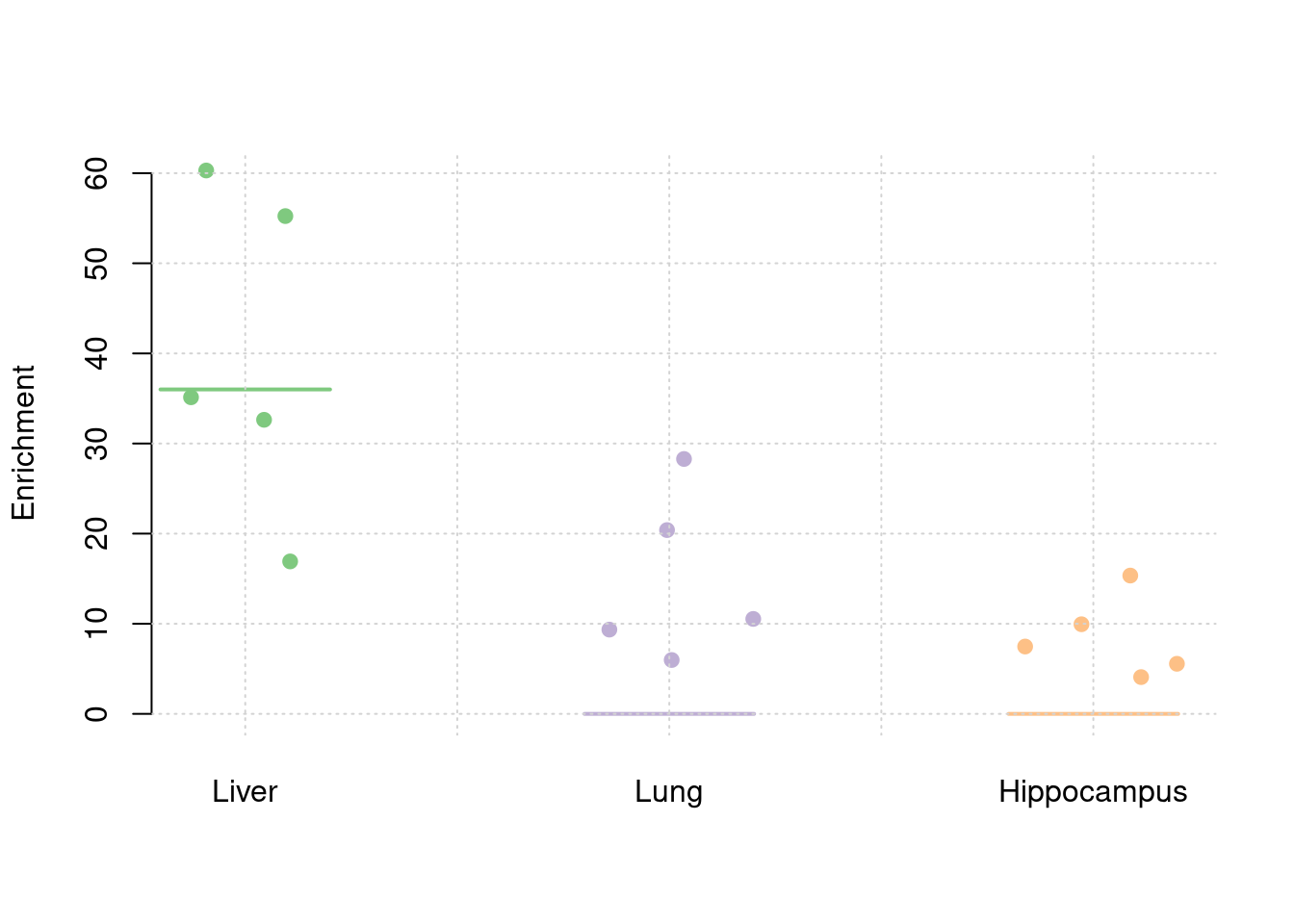
y1 <- results_df$pve_weight1
y2 <- results_df$pve_weight2
y3 <- results_df$pve_weight3
truth <- rbind(c(1,0.03),c(2,0),c(3,0))
est <- rbind(cbind(1,y1),cbind(2,y2),cbind(3,y3))
plot_par(truth,est,xlabels = c("Liver","Lung","Hippocampus"),ylim=c(0,0.05),ylab="PVE")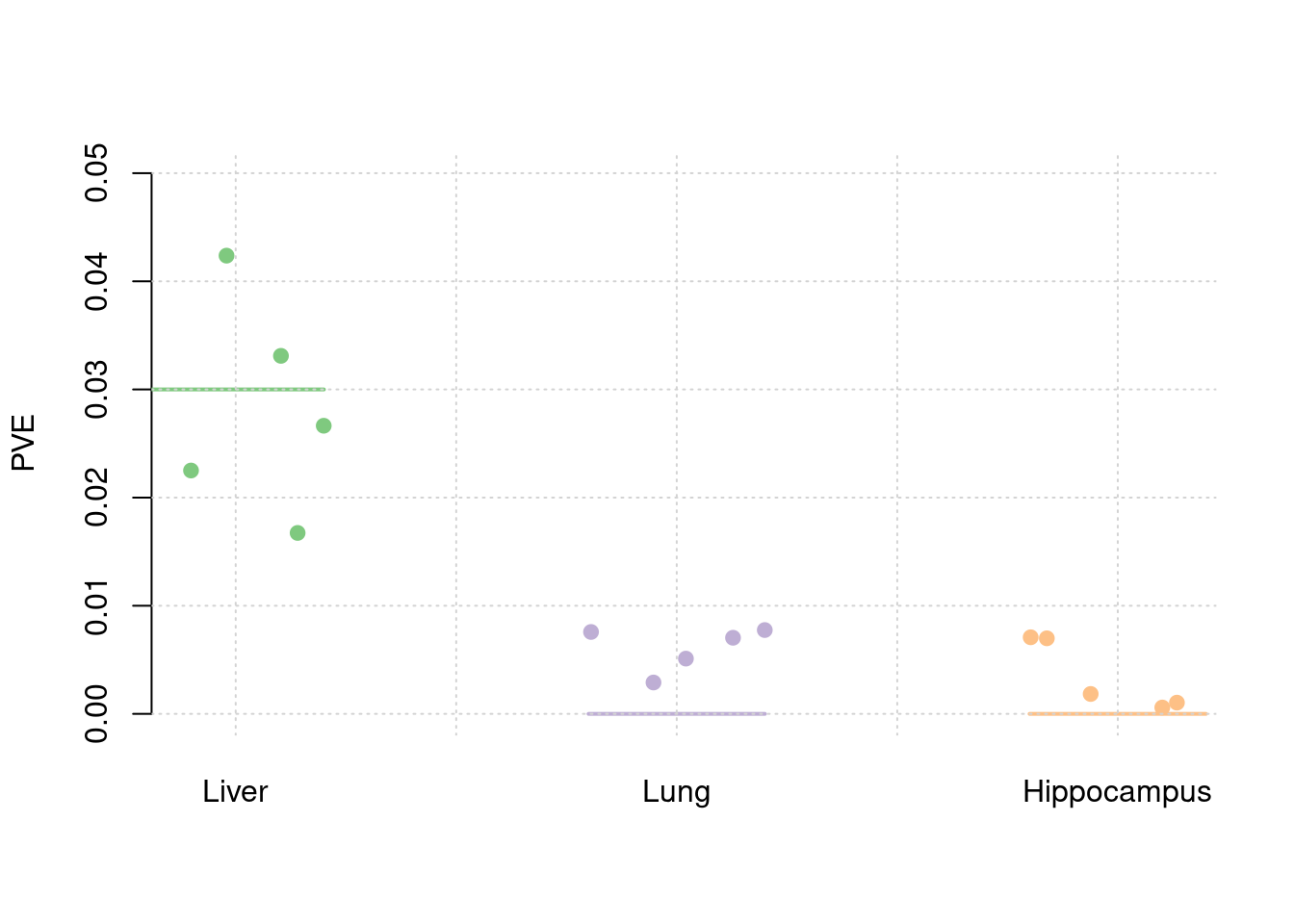
Individual tissue analyses
For the cTWAS analysis, each tissue was analyzed individually and the results were combined.
results_dir <- "/project2/xinhe/shengqian/cTWAS/cTWAS_simulation/simulation_uncorrelated_drop_merge/"
runtag = "ukb-s80.45-3_uncorr"
configtag <- 2
simutags <- paste(5, 1:5, sep = "-")
thin <- 0.1
sample_size <- 45000
PIP_threshold <- 0.8results_df <- get_sim_ind_res(results_dir,runtag,configtag,simutags,thin,sample_size,PIP_threshold) simutag n_detected_weight1 n_detected_in_causal_weight1
1 5-1 16 16
2 5-2 13 12
3 5-3 21 19
4 5-4 14 13
5 5-5 8 6[1] 0.9166667 simutag n_detected_weight2 n_detected_in_causal_weight2
1 5-1 2 2
2 5-2 6 4
3 5-3 9 5
4 5-4 5 3
5 5-5 4 2[1] 0.6153846 simutag n_detected_weight3 n_detected_in_causal_weight3
1 5-1 6 4
2 5-2 3 3
3 5-3 1 1
4 5-4 7 4
5 5-5 1 1[1] 0.7222222 simutag n_causal_combined n_detected_combined n_detected_in_causal_combined
1 5-1 68 18 16
2 5-2 63 15 12
3 5-3 78 25 19
4 5-4 66 19 13
5 5-5 54 10 6[1] 0.7586207Figure
For the cTWAS analysis, each tissue was analyzed individually and the results were combined. 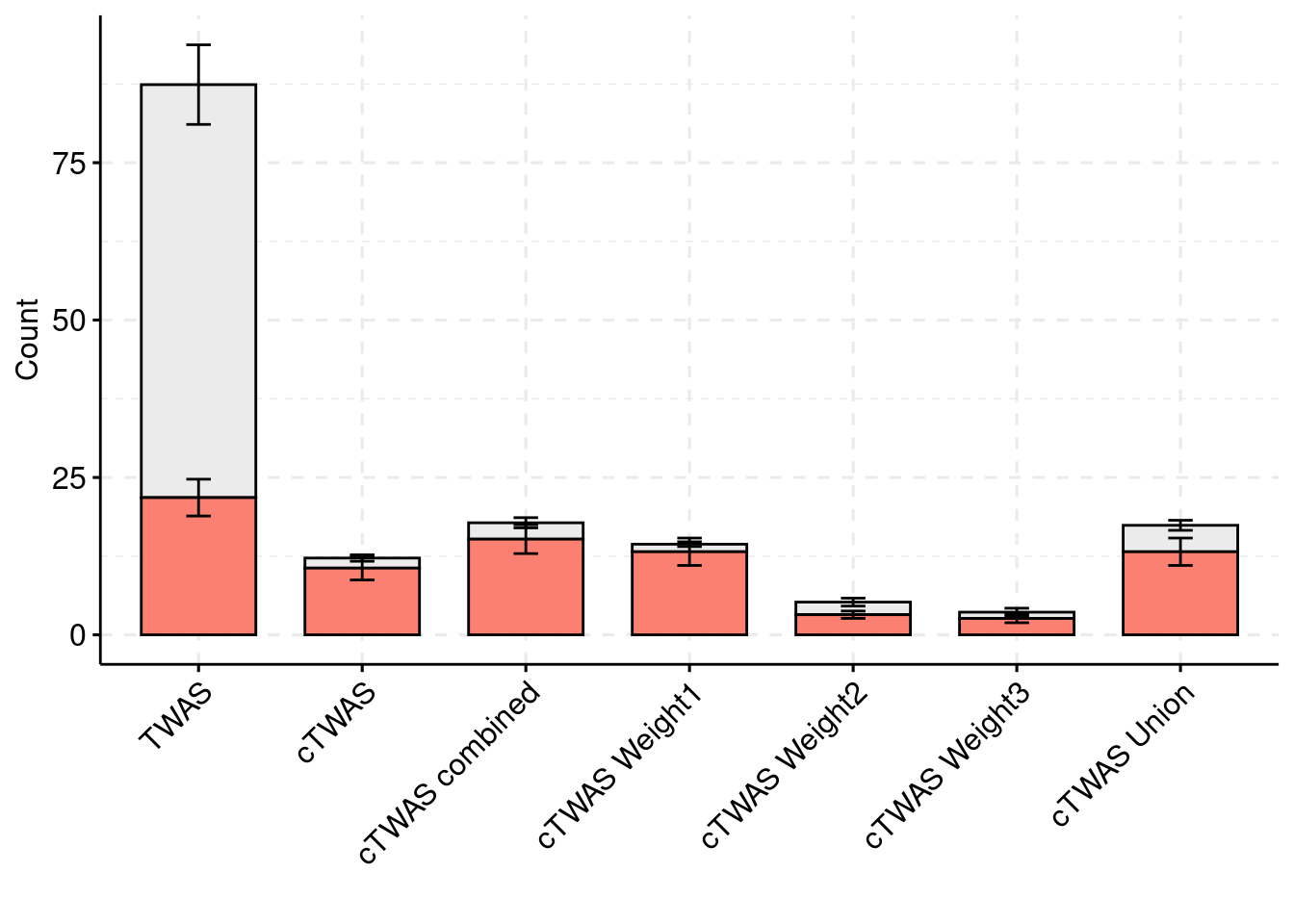
sessionInfo()R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggpubr_0.6.0 ggplot2_3.4.0 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] tidyselect_1.2.0 xfun_0.35 bslib_0.4.1 purrr_1.0.2
[5] carData_3.0-4 colorspace_2.0-3 vctrs_0.6.3 generics_0.1.3
[9] htmltools_0.5.4 yaml_2.3.6 utf8_1.2.2 rlang_1.1.1
[13] jquerylib_0.1.4 later_1.3.0 pillar_1.8.1 glue_1.6.2
[17] withr_2.5.0 DBI_1.1.3 lifecycle_1.0.3 stringr_1.5.0
[21] ggsignif_0.6.4 munsell_0.5.0 gtable_0.3.1 evaluate_0.19
[25] labeling_0.4.2 knitr_1.41 callr_3.7.3 fastmap_1.1.0
[29] httpuv_1.6.7 ps_1.7.2 fansi_1.0.3 highr_0.9
[33] broom_1.0.2 Rcpp_1.0.9 backports_1.2.1 promises_1.2.0.1
[37] scales_1.2.1 cachem_1.0.6 jsonlite_1.8.4 abind_1.4-5
[41] farver_2.1.0 fs_1.5.2 digest_0.6.31 stringi_1.7.8
[45] rstatix_0.7.2 processx_3.8.0 dplyr_1.0.10 getPass_0.2-2
[49] rprojroot_2.0.3 grid_4.1.0 cli_3.6.1 tools_4.1.0
[53] magrittr_2.0.3 sass_0.4.4 tibble_3.1.8 car_3.1-1
[57] tidyr_1.3.0 whisker_0.4.1 pkgconfig_2.0.3 data.table_1.14.6
[61] assertthat_0.2.1 rmarkdown_2.19 httr_1.4.4 rstudioapi_0.14
[65] R6_2.5.1 git2r_0.30.1 compiler_4.1.0