WhiteBlood - WholeBlood Traits Jointly
Sheng Qian
2023-2-1
Last updated: 2023-02-15
Checks: 5 2
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config_b38.R | code/ctwas_config_b38.R |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/mashr_sqtl/sqtl/mashr/mashr_Whole_Blood_Splicing_mapping.RData | data/mashr_sqtl/sqtl/mashr/mashr_Whole_Blood_Splicing_mapping.RData |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/mqtl/WholeBlood.db | data/mqtl/WholeBlood.db |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/G_list.RData | data/G_list.RData |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/locus_plot.R | code/locus_plot.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 1c8d70c. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .ipynb_checkpoints/
Untracked files:
Untracked: Proposal plots.R
Untracked: RGS14.pdf
Untracked: RNF186.pdf
Untracked: SCZ_annotation.xlsx
Untracked: SLC8B1.pdf
Untracked: analysis/.ipynb_checkpoints/
Untracked: cache/
Untracked: code/.ipynb_checkpoints/
Untracked: data/.ipynb_checkpoints/
Untracked: data/FUMA_output/
Untracked: data/GO_Terms/
Untracked: data/GTEx_Analysis_v8_eQTL.tar
Untracked: data/G_list.RData
Untracked: data/IBD_ME/
Untracked: data/LDL/
Untracked: data/LDL_E_S/
Untracked: data/LDL_M/
Untracked: data/LDL_S/
Untracked: data/PGC3_SCZ_wave3_public.v2.tsv
Untracked: data/SCZ/
Untracked: data/SCZ_2018/
Untracked: data/SCZ_2018_S/
Untracked: data/SCZ_2020/
Untracked: data/SCZ_S/
Untracked: data/Supplementary Table 15 - MAGMA.xlsx
Untracked: data/Supplementary Table 20 - Prioritised Genes.xlsx
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/WhiteBlood_E/
Untracked: data/WhiteBlood_E_M/
Untracked: data/WhiteBlood_E_S_M/
Untracked: data/WhiteBlood_M/
Untracked: data/cpg_annot.RData
Untracked: data/eqtl/
Untracked: data/gencode.v26.GRCh38.genes.gtf
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/gwas_sumstats/
Untracked: data/magma.genes.out
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/mqtl/
Untracked: data/notes.txt
Untracked: data/scz_2018.RDS
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: submit.sh
Untracked: temp_LDR/
Unstaged changes:
Deleted: analysis/Atrial_Fibrillation_Heart_Atrial_Appendage.Rmd
Deleted: analysis/Atrial_Fibrillation_Heart_Left_Ventricle.Rmd
Deleted: analysis/Autism_Brain_Amygdala.Rmd
Deleted: analysis/Autism_Brain_Anterior_cingulate_cortex_BA24.Rmd
Deleted: analysis/Autism_Brain_Caudate_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Cerebellar_Hemisphere.Rmd
Deleted: analysis/Autism_Brain_Cerebellum.Rmd
Deleted: analysis/Autism_Brain_Cortex.Rmd
Deleted: analysis/Autism_Brain_Frontal_Cortex_BA9.Rmd
Deleted: analysis/Autism_Brain_Hippocampus.Rmd
Deleted: analysis/Autism_Brain_Hypothalamus.Rmd
Deleted: analysis/Autism_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Putamen_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Spinal_cord_cervical_c-1.Rmd
Deleted: analysis/Autism_Brain_Substantia_nigra.Rmd
Deleted: analysis/BMI_Brain_Amygdala.Rmd
Deleted: analysis/BMI_Brain_Amygdala_S.Rmd
Deleted: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24.Rmd
Deleted: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24_S.Rmd
Deleted: analysis/BMI_Brain_Caudate_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Caudate_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Cerebellar_Hemisphere.Rmd
Deleted: analysis/BMI_Brain_Cerebellar_Hemisphere_S.Rmd
Deleted: analysis/BMI_Brain_Cerebellum.Rmd
Deleted: analysis/BMI_Brain_Cerebellum_S.Rmd
Deleted: analysis/BMI_Brain_Cortex.Rmd
Deleted: analysis/BMI_Brain_Cortex_S.Rmd
Deleted: analysis/BMI_Brain_Frontal_Cortex_BA9.Rmd
Deleted: analysis/BMI_Brain_Frontal_Cortex_BA9_S.Rmd
Deleted: analysis/BMI_Brain_Hippocampus.Rmd
Deleted: analysis/BMI_Brain_Hippocampus_S.Rmd
Deleted: analysis/BMI_Brain_Hypothalamus.Rmd
Deleted: analysis/BMI_Brain_Hypothalamus_S.Rmd
Deleted: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Putamen_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Putamen_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Spinal_cord_cervical_c-1.Rmd
Deleted: analysis/BMI_Brain_Spinal_cord_cervical_c-1_S.Rmd
Deleted: analysis/BMI_Brain_Substantia_nigra.Rmd
Deleted: analysis/BMI_Brain_Substantia_nigra_S.Rmd
Deleted: analysis/BMI_S_results.Rmd
Deleted: analysis/Glucose_Adipose_Subcutaneous.Rmd
Deleted: analysis/Glucose_Adipose_Visceral_Omentum.Rmd
Modified: analysis/WhiteBlood_WholeBlood_E.Rmd
Modified: analysis/WhiteBlood_WholeBlood_E_M.Rmd
Modified: analysis/WhiteBlood_WholeBlood_E_S_M.Rmd
Modified: analysis/WhiteBlood_WholeBlood_M.Rmd
Deleted: code/White_Blood_M_out/White_Blood_BreastMammary.err
Deleted: code/White_Blood_M_out/White_Blood_BreastMammary.out
Deleted: code/White_Blood_M_out/White_Blood_ColonTransverse.err
Deleted: code/White_Blood_M_out/White_Blood_ColonTransverse.out
Deleted: code/White_Blood_M_out/White_Blood_KidneyCortex.err
Deleted: code/White_Blood_M_out/White_Blood_KidneyCortex.out
Deleted: code/White_Blood_M_out/White_Blood_Lung.err
Deleted: code/White_Blood_M_out/White_Blood_Lung.out
Deleted: code/White_Blood_M_out/White_Blood_MuscleSkeletal.err
Deleted: code/White_Blood_M_out/White_Blood_MuscleSkeletal.out
Deleted: code/White_Blood_M_out/White_Blood_Ovary.err
Deleted: code/White_Blood_M_out/White_Blood_Ovary.out
Deleted: code/White_Blood_M_out/White_Blood_Prostate.err
Deleted: code/White_Blood_M_out/White_Blood_Prostate.out
Deleted: code/White_Blood_M_out/White_Blood_Testis.err
Deleted: code/White_Blood_M_out/White_Blood_Testis.out
Deleted: code/White_Blood_M_out/White_Blood_WholeBlood.err
Deleted: code/White_Blood_M_out/White_Blood_WholeBlood.out
Deleted: code/run_IBD_ctwas_rss_LDR_ME.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/WhiteBlood_WholeBlood_E_S_M.Rmd) and HTML (docs/WhiteBlood_WholeBlood_E_S_M.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 213f0e4 | sq-96 | 2023-02-15 | update |
| html | 213f0e4 | sq-96 | 2023-02-15 | update |
| Rmd | 0e327ac | sq-96 | 2023-02-07 | update |
| html | 0e327ac | sq-96 | 2023-02-07 | update |
| Rmd | 1b50100 | sq-96 | 2023-02-07 | update |
| html | 1b50100 | sq-96 | 2023-02-07 | update |
| Rmd | 5b2c6c6 | sq-96 | 2023-02-07 | update |
| Rmd | f49450b | sq-96 | 2023-02-04 | update |
| html | f49450b | sq-96 | 2023-02-04 | update |
| Rmd | da7a0f7 | sq-96 | 2023-02-03 | update |
| html | da7a0f7 | sq-96 | 2023-02-03 | update |
| Rmd | ba657aa | sq-96 | 2023-02-03 | update |
| html | ba657aa | sq-96 | 2023-02-03 | update |
| Rmd | fa89b37 | sq-96 | 2023-02-03 | update |
| html | fa89b37 | sq-96 | 2023-02-03 | update |
| Rmd | 6286c34 | sq-96 | 2023-02-02 | update |
| html | 6286c34 | sq-96 | 2023-02-02 | update |
| Rmd | a762568 | sq-96 | 2023-02-02 | update |
| html | a762568 | sq-96 | 2023-02-02 | update |
| Rmd | c5aaf96 | sq-96 | 2023-02-02 | update |
| html | c5aaf96 | sq-96 | 2023-02-02 | update |
| Rmd | 9b01dad | sq-96 | 2023-02-01 | update |
analysis_id <- params$analysis_id
trait_id <- params$trait_id
weight <- params$weight
results_dir <- paste0("/project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/", trait_id, "/", weight)
source("/project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config_b38.R")
options(digits = 4)Load ctwas results
Check convergence of parameters
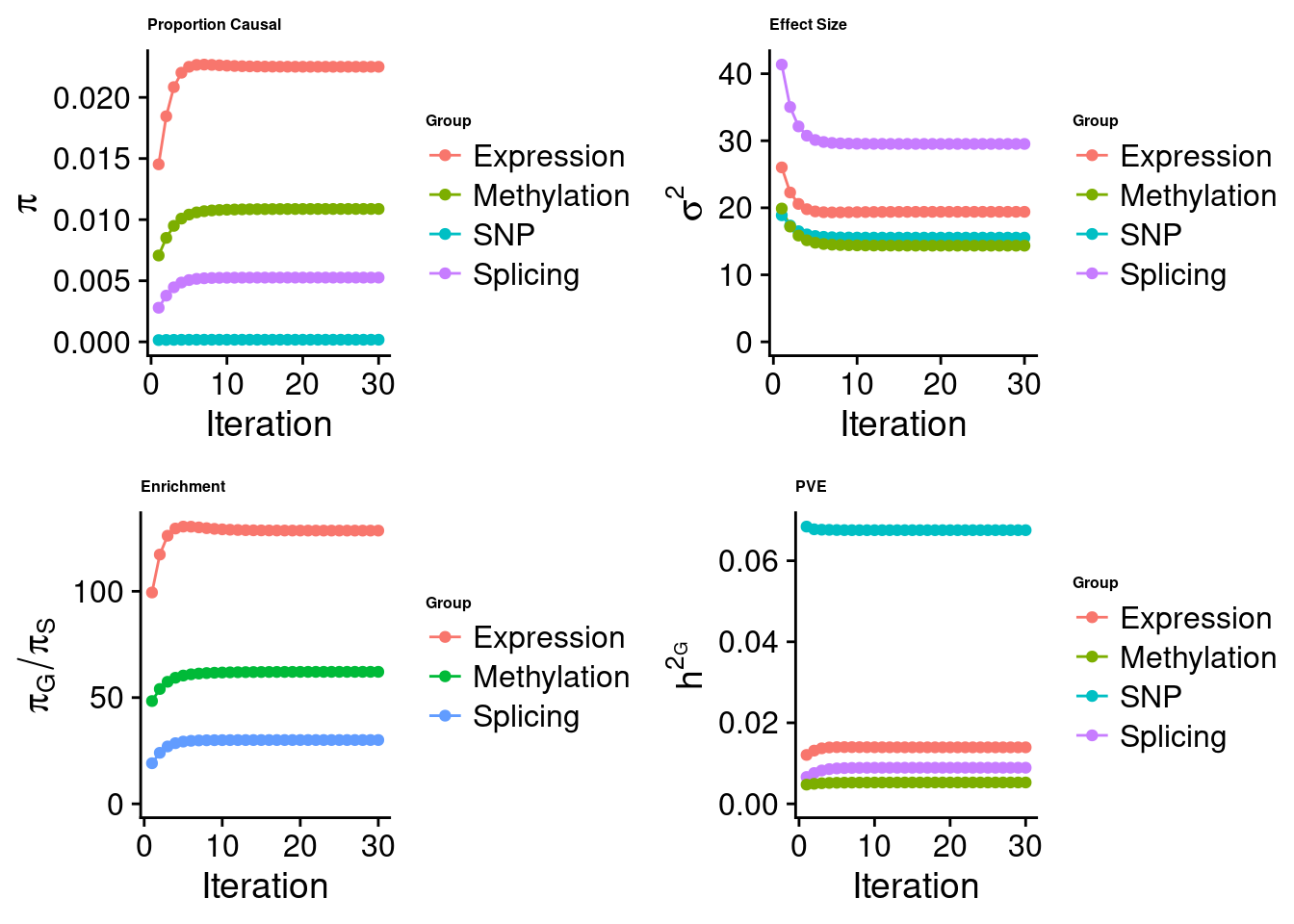
#estimated group prior
estimated_group_prior <- estimated_group_prior_all[,ncol(group_prior_rec)]
print(estimated_group_prior) SNP Whole_Blood Whole_Blood_Splicing
0.0001751 0.0225055 0.0052580
Whole_Blood_Methylation
0.0108737 #estimated group prior variance
estimated_group_prior_var <- estimated_group_prior_var_all[,ncol(group_prior_var_rec)]
print(estimated_group_prior_var) SNP Whole_Blood Whole_Blood_Splicing
15.55 19.40 29.53
Whole_Blood_Methylation
14.35 #estimated enrichment
estimated_enrichment <- estimated_enrichment_all[ncol(group_prior_var_rec)]
print(estimated_enrichment)[1] 61.79#report sample size
print(sample_size)[1] 350470#report group size
print(group_size) SNP Whole_Blood Whole_Blood_Splicing
8696600 11198 20134
Whole_Blood_Methylation
11858 #estimated group PVE
estimated_group_pve <- estimated_group_pve_all[,ncol(group_prior_rec)]
print(estimated_group_pve) SNP Whole_Blood Whole_Blood_Splicing
0.067547 0.013950 0.008919
Whole_Blood_Methylation
0.005278 #total PVE
sum(estimated_group_pve)[1] 0.09569#attributable PVE
estimated_group_pve/sum(estimated_group_pve) SNP Whole_Blood Whole_Blood_Splicing
0.70586 0.14578 0.09320
Whole_Blood_Methylation
0.05515 Top expression/intron/CpG units
genename gene_id susie_pip group region_tag
39936 FES ENSG00000182511.11 1.0000 Expression 15_43
43125 YBEY intron_21_46287123_46296162 1.0000 Splicing 21_24
38047 TAGAP ENSG00000164691.16 1.0000 Expression 6_103
36651 ARHGAP15 ENSG00000075884.13 1.0000 Expression 2_85
41903 CNN2 intron_19_1032696_1036130 1.0000 Splicing 19_2
36665 BAZ2B ENSG00000123636.17 0.9999 Expression 2_96
38535 VLDLR ENSG00000147852.15 0.9999 Expression 9_3
38156 CREB5 ENSG00000146592.16 0.9991 Expression 7_24
37171 MED12L ENSG00000144893.12 0.9981 Expression 3_93
35946 LAPTM5 ENSG00000162511.7 0.9981 Expression 1_20
37527 SLC22A4 ENSG00000197208.5 0.9966 Expression 5_79
37106 ATXN7 ENSG00000163635.17 0.9965 Expression 3_43
39549 FLT3 intron_13_28024943_28027088 0.9949 Splicing 13_7
36279 CNIH4 ENSG00000143771.11 0.9948 Expression 1_114
39447 ALDH2 ENSG00000111275.12 0.9941 Expression 12_67
39604 LAMP1 ENSG00000185896.10 0.9931 Expression 13_62
36089 SLC25A24 ENSG00000085491.15 0.9924 Expression 1_67
43166 HDHD5 ENSG00000069998.12 0.9912 Expression 22_1
41827 TTC39C ENSG00000168234.12 0.9872 Expression 18_12
41159 YPEL2 ENSG00000175155.8 0.9846 Expression 17_34
38704 LIPA ENSG00000107798.17 0.9839 Expression 10_57
41100 KIF18B ENSG00000186185.13 0.9837 Expression 17_26
39339 ACVRL1 cg21236262 0.9824 Methylation 12_33
38973 PTPRJ ENSG00000149177.12 0.9824 Expression 11_29
37291 ARAP2 ENSG00000047365.11 0.9783 Expression 4_30
37629 CPEB4 ENSG00000113742.12 0.9741 Expression 5_104
40700 MYO1C cg02622416 0.9736 Methylation 17_2
40445 ZFPM1 ENSG00000179588.8 0.9711 Expression 16_54
38575 OSTF1 ENSG00000134996.11 0.9691 Expression 9_35
39588 KLF12 ENSG00000118922.17 0.9685 Expression 13_36
36440 ITSN2 intron_2_24209221_24209818 0.9663 Splicing 2_14
37693 ATXN1 ENSG00000124788.17 0.9616 Expression 6_13
38633 NEK6 ENSG00000119408.16 0.9567 Expression 9_64
36934 UQCRC1 ENSG00000010256.10 0.9555 Expression 3_34
40367 GLG1 ENSG00000090863.11 0.9553 Expression 16_40
37233 ADD1 ENSG00000087274.16 0.9550 Expression 4_4
37348 TET2 ENSG00000168769.13 0.9535 Expression 4_69
36222 SELL ENSG00000188404.8 0.9505 Expression 1_83
36005 CITED4 ENSG00000179862.6 0.9505 Expression 1_25
37276 TBC1D14 ENSG00000132405.18 0.9442 Expression 4_8
37664 CANX intron_5_179699102_179705679 0.9434 Splicing 5_108
36622 HS6ST1 ENSG00000136720.6 0.9385 Expression 2_75
36670 CD302 ENSG00000241399.6 0.9303 Expression 2_96
39154 CD6 cg27098804 0.9266 Methylation 11_34
36067 CDC14A ENSG00000079335.18 0.9259 Expression 1_61
37398 DDX60L ENSG00000181381.13 0.9238 Expression 4_109
42503 ATP13A1 ENSG00000105726.16 0.9228 Expression 19_15
42362 MYO9B ENSG00000099331.13 0.9165 Expression 19_14
40361 ITGAL cg24033122 0.9158 Methylation 16_24
42233 AP1M2 ENSG00000129354.11 0.9144 Expression 19_9
36881 CCR8 ENSG00000179934.6 0.9073 Expression 3_28
37134 PAQR9 ENSG00000188582.8 0.9002 Expression 3_87
37511 TNFAIP8 ENSG00000145779.7 0.8984 Expression 5_72
37439 RAI14 ENSG00000039560.13 0.8925 Expression 5_23
38269 CPSF4 ENSG00000160917.14 0.8870 Expression 7_61
37608 ADAM19 ENSG00000135074.15 0.8862 Expression 5_93
36265 NCF2 intron_1_183570839_183573185 0.8821 Splicing 1_91
41207 SLC9A3R1 ENSG00000109062.11 0.8776 Expression 17_42
37185 ABCC5 ENSG00000114770.16 0.8707 Expression 3_112
41423 UBE2O ENSG00000175931.12 0.8706 Expression 17_43
37312 REST ENSG00000084093.16 0.8678 Expression 4_41
39271 CERS5 intron_12_50134068_50134546 0.8677 Splicing 12_33
42844 UBOX5 ENSG00000185019.16 0.8644 Expression 20_5
36462 DTNB ENSG00000138101.18 0.8556 Expression 2_15
37475 ZBED3 ENSG00000132846.5 0.8554 Expression 5_45
38383 ACHE ENSG00000087085.13 0.8461 Expression 7_62
38011 L3MBTL3 ENSG00000198945.7 0.8426 Expression 6_86
37418 NPR3 ENSG00000113389.15 0.8420 Expression 5_22
37369 LRBA intron_4_150278004_150282450 0.8388 Splicing 4_98
37420 MTMR12 ENSG00000150712.10 0.8381 Expression 5_22
36185 RCSD1 ENSG00000198771.10 0.8329 Expression 1_82
41409 JMJD6 ENSG00000070495.14 0.8306 Expression 17_43
37118 PXYLP1 ENSG00000155893.12 0.8284 Expression 3_86
39582 KLF5 ENSG00000102554.13 0.8242 Expression 13_35
41886 ELANE ENSG00000197561.6 0.8125 Expression 19_2
38127 JAZF1 ENSG00000153814.11 0.8109 Expression 7_23
37994 HSF2 ENSG00000025156.12 0.8108 Expression 6_82
39810 ZFP36L1 ENSG00000185650.9 0.8106 Expression 14_33
42600 LSM4 cg15796753 0.8058 Methylation 19_15
38758 RBP4 ENSG00000138207.13 0.8035 Expression 10_59Top genes by expression/splicing/methylation pip
genename susie_pip group
13036 MYO1G 1.3514 Splicing
18333 NINJ2 1.1701 Methylation
15394 TSPAN32 1.1599 Splicing
16152 AMZ1 1.0639 Methylation
12354 ITSN2 1.0254 Splicing
14458 SIRPB1 1.0210 Splicing
18816 PTPRN2 1.0060 Methylation
12655 LYZ 1.0001 Splicing
11001 CNN2 1.0000 Splicing
2988 FES 1.0000 Expression
15750 YBEY 1.0000 Splicing
8211 TAGAP 1.0000 Expression
536 ARHGAP15 1.0000 Expression
812 BAZ2B 0.9999 Expression
9260 VLDLR 0.9999 Expression
13952 RAB34 0.9995 Splicing
1935 CREB5 0.9991 Expression
17144 ELK3 0.9989 Methylation
4774 MED12L 0.9981 Expression
4334 LAPTM5 0.9981 Expression
7574 SLC22A4 0.9966 Expression
750 ATXN7 0.9965 Expression
11772 FLT3 0.9955 Splicing
1761 CNIH4 0.9948 Expression
315 ALDH2 0.9941 Expression
10662 CANX 0.9939 Splicing
4324 LAMP1 0.9931 Expression
7591 SLC25A24 0.9924 Expression
3601 HDHD5 0.9912 Expression
19344 SPTLC2 0.9895 Methylation
8964 TTC39C 0.9872 Expression
9441 YPEL2 0.9846 Expression
4431 LIPA 0.9839 Expression
4188 KIF18B 0.9837 Expression
16046 ACVRL1 0.9824 Methylation
6614 PTPRJ 0.9824 Expression
15239 TNFRSF1A 0.9794 Splicing
517 ARAP2 0.9783 Expression
1896 CPEB4 0.9741 Expression
18244 MYO1C 0.9736 Methylation
9547 ZFPM1 0.9711 Expression
19006 RRBP1 0.9698 Methylation
5750 OSTF1 0.9691 Expression
4219 KLF12 0.9685 Expression
744 ATXN1 0.9616 Expression
5376 NEK6 0.9567 Expression
9163 UQCRC1 0.9555 Expression
3293 GLG1 0.9553 Expression
181 ADD1 0.9550 Expression
8342 TET2 0.9535 Expression
19040 SBNO2 0.9509 Methylation
7349 SELL 0.9505 Expression
1658 CITED4 0.9505 Expression
8245 TBC1D14 0.9442 Expression
13092 NCF2 0.9422 Splicing
3753 HS6ST1 0.9385 Expression
15137 TMCC2 0.9337 Splicing
1386 CD302 0.9303 Expression
16612 CD6 0.9266 Methylation
1422 CDC14A 0.9259 Expression
2212 DDX60L 0.9238 Expression
706 ATP13A1 0.9228 Expression
5206 MYO9B 0.9165 Expression
17736 ITGAL 0.9161 Methylation
452 AP1M2 0.9144 Expression
1345 CCR8 0.9073 Expression
12807 MFSD13A 0.9073 Splicing
13846 PSD4 0.9016 Splicing
5828 PAQR9 0.9002 Expression
8661 TNFAIP8 0.8984 Expression
19844 ZBTB2 0.8967 Methylation
6744 RAI14 0.8925 Expression
1912 CPSF4 0.8870 Expression
154 ADAM19 0.8862 Expression
15682 WDFY2 0.8849 Splicing
7716 SLC9A3R1 0.8776 Expression
36 ABCC5 0.8707 Expression
9069 UBE2O 0.8706 Expression
6880 REST 0.8678 Expression
10886 CERS5 0.8677 Splicing
9089 UBOX5 0.8644 Expression
13614 PLCB2 0.8623 Splicing
17282 FBRSL1 0.8596 Methylation
2424 DTNB 0.8556 Expression
9457 ZBED3 0.8554 Expression
91 ACHE 0.8461 Expression
4310 L3MBTL3 0.8426 Expression
5534 NPR3 0.8420 Expression
19723 UBE2I 0.8420 Methylation
12594 LRBA 0.8388 Splicing
5123 MTMR12 0.8381 Expression
6854 RCSD1 0.8329 Expression
4077 JMJD6 0.8306 Expression
6643 PXYLP1 0.8284 Expression
4223 KLF5 0.8242 Expression
10712 CCDC125 0.8207 Splicing
2581 ELANE 0.8125 Expression
4074 JAZF1 0.8109 Expression
3770 HSF2 0.8108 Expression
9537 ZFP36L1 0.8106 Expression
17992 LSM4 0.8058 Methylation
16212 APOLD1 0.8042 Methylation
6832 RBP4 0.8035 ExpressionTop genes by combined PIP
genename combined_pip expression_pip splicing_pip methylation_pip
11761 YPEL2 1.753 0.985 0.000 0.768
5450 LAPTM5 1.744 0.998 0.002 0.744
665 ARHGAP15 1.723 1.000 0.680 0.042
6495 MYO1G 1.433 0.082 1.351 0.000
9593 SLC45A4 1.313 0.786 0.509 0.018
2770 DDX60L 1.245 0.924 0.322 0.000
10814 TNFAIP8 1.238 0.898 0.120 0.219
4508 HDHD5 1.217 0.991 0.226 0.000
6779 NINJ2 1.209 0.033 0.006 1.170
5772 LYZ 1.183 0.182 1.000 0.000
9946 SPTLC2 1.176 0.187 0.000 0.989
11126 TSPAN32 1.163 0.004 1.160 0.000
8908 RRBP1 1.129 0.066 0.093 0.970
9412 SLC12A7 1.093 0.416 0.011 0.666
921 ATXN1 1.086 0.962 0.058 0.067
98 ACAP1 1.083 0.349 0.015 0.719
5067 ITSN2 1.071 0.033 1.025 0.013
452 AMZ1 1.064 0.000 0.000 1.064
9375 SIRPB1 1.058 0.037 1.021 0.000
4984 IQGAP2 1.056 0.443 0.589 0.024
228 ADD1 1.055 0.955 0.100 0.000
7999 PPP5C 1.050 0.436 0.004 0.610
11848 ZDHHC18 1.048 0.009 0.590 0.450
6808 NLRC5 1.047 0.676 0.074 0.297
3336 EOMES 1.043 0.645 0.009 0.389
3258 ELK3 1.041 0.031 0.012 0.999
927 ATXN7 1.040 0.997 0.043 0.000
3663 FBRSL1 1.039 0.063 0.117 0.860
10828 TNFRSF1A 1.037 0.016 0.979 0.042
8263 PTPRJ 1.035 0.982 0.015 0.038
10315 TBC1D14 1.031 0.944 0.062 0.025
1699 CD101 1.028 0.702 0.006 0.320
2203 CNIH4 1.018 0.995 0.023 0.000
5984 MED12L 1.015 0.998 0.000 0.017
384 ALDH2 1.014 0.994 0.020 0.000
3755 FES 1.012 1.000 0.012 0.000
5296 KLF12 1.007 0.968 0.038 0.000
8267 PTPRN2 1.006 0.000 0.000 1.006
3398 ERICH1 1.004 0.046 0.203 0.754
11531 VLDLR 1.004 1.000 0.004 0.000
2424 CREB5 1.001 0.999 0.002 0.000
8363 RAB34 1.001 0.001 1.000 0.000
10273 TAGAP 1.001 1.000 0.001 0.000
1010 BAZ2B 1.000 1.000 0.001 0.000
2206 CNN2 1.000 0.000 1.000 0.000
2377 CPEB4 1.000 0.974 0.000 0.025
6491 MYO1C 1.000 0.026 0.000 0.974
11741 YBEY 1.000 0.000 1.000 0.000
639 ARAP2 0.999 0.978 0.021 0.000
9455 SLC22A4 0.998 0.997 0.002 0.000
9476 SLC25A24 0.997 0.992 0.004 0.000
3829 FLT3 0.995 0.000 0.995 0.000
1445 CANX 0.994 0.000 0.994 0.000
5437 LAMP1 0.993 0.993 0.000 0.000
7197 OSTF1 0.992 0.969 0.023 0.000
5567 LIPA 0.991 0.984 0.006 0.001
8810 RORC 0.991 0.776 0.000 0.216
1732 CD33 0.990 0.039 0.767 0.184
11173 TTC39C 0.988 0.987 0.001 0.000
5253 KIF18B 0.984 0.984 0.000 0.000
7704 PLCB2 0.984 0.121 0.862 0.000
175 ACVRL1 0.982 0.000 0.000 0.982
10605 TMCC2 0.978 0.044 0.934 0.000
10956 TRAF3IP3 0.978 0.686 0.292 0.000
11888 ZFPM1 0.973 0.971 0.001 0.000
11295 UBE2I 0.968 0.067 0.059 0.842
6709 NEK6 0.966 0.957 0.004 0.006
7054 NUP88 0.965 0.172 0.767 0.026
3055 DTNB 0.962 0.856 0.005 0.102
4139 GLG1 0.958 0.955 0.003 0.000
9609 SLC5A11 0.958 0.302 0.000 0.656
11326 UBOX5 0.957 0.864 0.092 0.000
10429 TET2 0.956 0.953 0.003 0.000
11413 UQCRC1 0.956 0.956 0.000 0.000
4698 HS6ST1 0.955 0.939 0.000 0.016
9172 SELL 0.955 0.951 0.005 0.000
9046 SBNO2 0.952 0.000 0.001 0.951
2078 CITED4 0.951 0.951 0.000 0.000
6162 MLX 0.949 0.735 0.214 0.000
6611 NCF2 0.942 0.000 0.942 0.000
11789 ZBTB2 0.936 0.040 0.000 0.897
4375 GSAP 0.932 0.630 0.262 0.040
1730 CD302 0.930 0.930 0.000 0.000
1750 CD6 0.928 0.000 0.001 0.927
1771 CDC14A 0.926 0.926 0.000 0.000
867 ATP13A1 0.925 0.923 0.002 0.000
1299 C20orf96 0.920 0.721 0.060 0.138
11614 WDFY2 0.918 0.000 0.885 0.034
46 ABCC5 0.917 0.871 0.019 0.027
6504 MYO9B 0.917 0.917 0.000 0.000
5036 ITGAL 0.916 0.000 0.000 0.916
562 AP1M2 0.914 0.914 0.000 0.000
5219 KIAA0040 0.914 0.067 0.085 0.762
8422 RAI14 0.913 0.892 0.000 0.021
1684 CCR8 0.907 0.907 0.000 0.000
6077 MFSD13A 0.907 0.000 0.907 0.000
6401 MTMR12 0.902 0.838 0.064 0.000
8152 PSD4 0.902 0.000 0.902 0.000
186 ADAM19 0.900 0.886 0.005 0.008
7296 PAQR9 0.900 0.900 0.000 0.000
9005 SAE1 0.894 0.622 0.272 0.000
2396 CPSF4 0.887 0.887 0.000 0.000
5841 MAP2K5 0.880 0.070 0.030 0.780
9639 SLC9A3R1 0.878 0.878 0.000 0.000
1733 CD36 0.877 0.563 0.314 0.000
5089 JAZF1 0.874 0.811 0.063 0.000
5661 LRRC25 0.873 0.735 0.137 0.000
7015 NUDT14 0.871 0.216 0.557 0.098
11302 UBE2O 0.871 0.871 0.001 0.000
5420 L3MBTL3 0.870 0.843 0.000 0.027
1945 CERS5 0.868 0.000 0.868 0.000
8596 REST 0.868 0.868 0.000 0.000
5632 LRBA 0.861 0.000 0.839 0.022
1626 CCDC9 0.858 0.667 0.191 0.000
2958 DNASE1 0.858 0.551 0.007 0.300
1547 CCDC125 0.855 0.034 0.821 0.000
11782 ZBED3 0.855 0.855 0.000 0.000
114 ACHE 0.851 0.846 0.005 0.000
8234 PTK2B 0.849 0.739 0.053 0.057
8566 RCSD1 0.846 0.833 0.008 0.005
8145 PRUNE2 0.843 0.799 0.006 0.038
6906 NPR3 0.842 0.842 0.000 0.000
759 ARRB2 0.836 0.281 0.554 0.000
5094 JMJD6 0.836 0.831 0.005 0.000
8300 PXYLP1 0.836 0.828 0.007 0.000
3263 ELMO1 0.835 0.702 0.046 0.086
624 APOLD1 0.833 0.029 0.000 0.804
5301 KLF5 0.824 0.824 0.000 0.000
5712 LSM4 0.818 0.011 0.001 0.806
1502 CATSPER2 0.815 0.792 0.000 0.023
3251 ELANE 0.813 0.813 0.000 0.000
8935 RSU1 0.812 0.037 0.270 0.505
4716 HSF2 0.811 0.811 0.000 0.000
11876 ZFP36L1 0.811 0.811 0.000 0.000
674 ARHGAP26 0.810 0.000 0.022 0.788
694 ARHGEF12 0.809 0.762 0.046 0.000
182 ADAM10 0.807 0.798 0.009 0.000
5893 MARK3 0.807 0.696 0.109 0.002
12115 ZNF516 0.807 0.792 0.004 0.011
8541 RBP4 0.804 0.804 0.000 0.000
region_tag
11761 17_34
5450 1_20
665 2_85
6495 7_33
9593 8_92
2770 4_109
10814 5_72
4508 22_1
6779 12_1
5772 12_42
9946 14_36
11126 11_2
8908 20_13
9412 5_2
921 6_13
98 17_6
5067 2_14
452 7_4
9375 20_2
4984 5_44
228 4_4
7999 19_32
11848 1_18
6808 16_31
3336 3_20
3258 12_57
927 3_43
3663 12_82
10828 12_7
8263 11_29
10315 4_8
1699 1_72
2203 1_114
5984 3_93
384 12_67
3755 15_43
5296 13_36
8267 7_98
3398 8_1
11531 9_3
2424 7_24
8363 17_18
10273 6_103
1010 2_96
2206 19_2
2377 5_104
6491 17_2
11741 21_24
639 4_30
9455 5_79
9476 1_67
3829 13_7
1445 5_108
5437 13_62
7197 9_35
5567 10_57
8810 1_74
1732 19_35
11173 18_12
5253 17_26
7704 15_14
175 12_33
10605 1_104
10956 1_106
11888 16_54
11295 16_2
6709 9_64
7054 17_5
3055 2_15
4139 16_40
9609 16_21
11326 20_5
10429 4_69
11413 3_34
4698 2_75
9172 1_83
9046 19_2
2078 1_25
6162 17_25
6611 1_91
11789 6_98
4375 7_49
1730 2_96
1750 11_34
1771 1_61
867 19_15
1299 20_1
11614 13_22
46 3_112
6504 19_14
5036 16_24
562 19_9
5219 1_86
8422 5_23
1684 3_28
6077 10_65
6401 5_22
8152 2_67
186 5_93
7296 3_87
9005 19_33
2396 7_61
5841 15_31
9639 17_42
1733 7_51
5089 7_23
5661 19_15
7015 14_55
11302 17_43
5420 6_86
1945 12_33
8596 4_41
5632 4_98
1626 19_33
2958 16_3
1547 5_42
11782 5_45
114 7_62
8234 8_27
8566 1_82
8145 9_36
6906 5_22
759 17_4
5094 17_43
8300 3_86
3263 7_27
624 12_12
5301 13_35
5712 19_15
1502 15_16
3251 19_2
8935 10_14
4716 6_82
11876 14_33
674 5_84
694 11_72
182 15_27
5893 14_54
12115 18_45
8541 10_59GO enrichment analysis for genes with PIP>0.5
#number of genes for gene set enrichment
length(genes)[1] 140Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"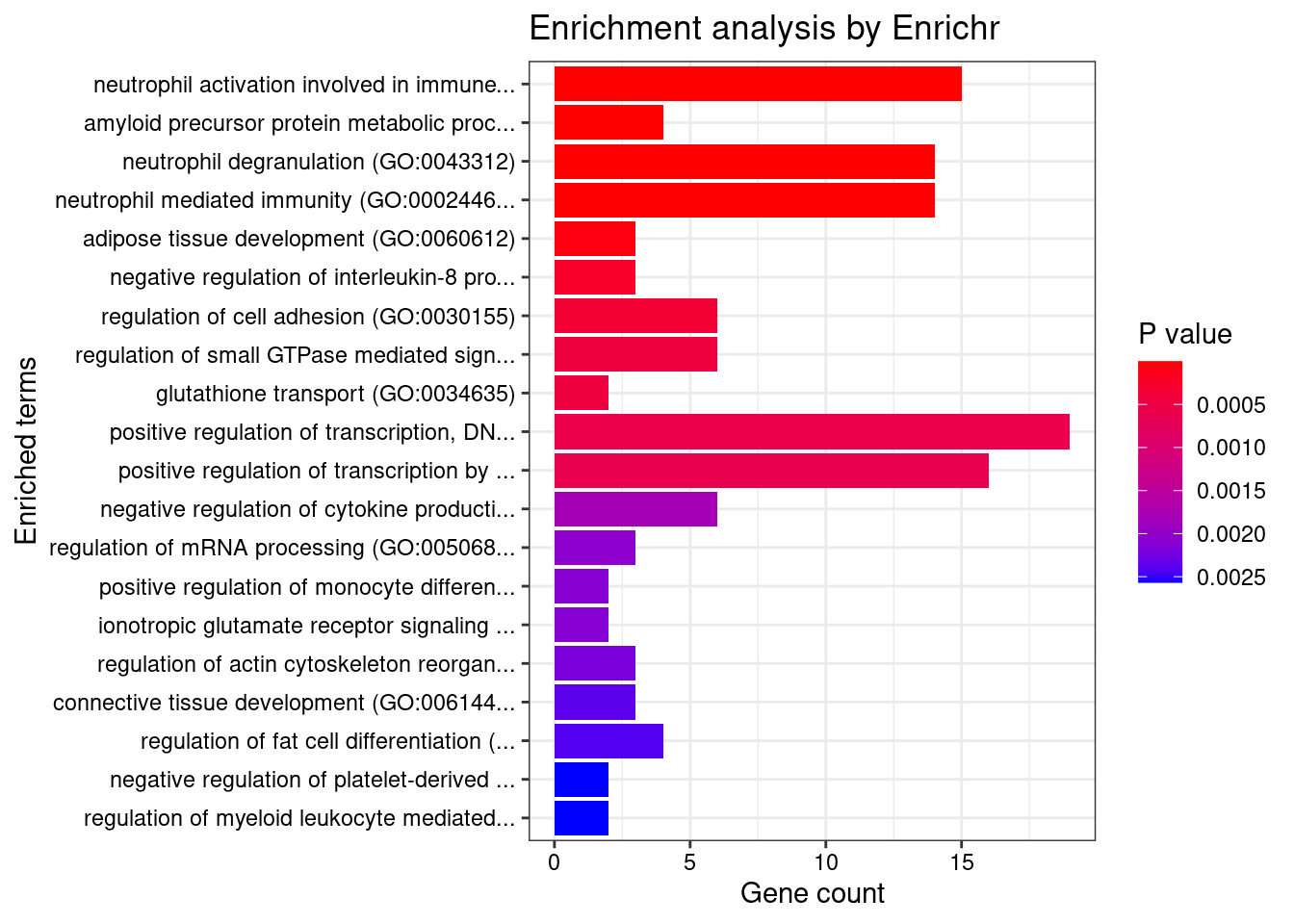
| Version | Author | Date |
|---|---|---|
| 1b50100 | sq-96 | 2023-02-07 |
Term Overlap
1 neutrophil activation involved in immune response (GO:0002283) 15/485
2 amyloid precursor protein metabolic process (GO:0042982) 4/18
3 neutrophil degranulation (GO:0043312) 14/481
4 neutrophil mediated immunity (GO:0002446) 14/488
5 adipose tissue development (GO:0060612) 3/12
Adjusted.P.value
1 0.002091
2 0.002795
3 0.002795
4 0.002795
5 0.018207
Genes
1 PTPRN2;ADAM10;PTPRJ;IQGAP2;LYZ;ITGAL;SIRPB1;CNN2;LAMP1;SELL;OSTF1;CD36;DNASE1;ELANE;CD33
2 ADAM19;ACHE;ADAM10;TMCC2
3 PTPRN2;ADAM10;PTPRJ;IQGAP2;LYZ;ITGAL;SIRPB1;CNN2;LAMP1;SELL;OSTF1;CD36;ELANE;CD33
4 PTPRN2;ADAM10;PTPRJ;IQGAP2;LYZ;ITGAL;SIRPB1;CNN2;LAMP1;SELL;OSTF1;CD36;ELANE;CD33
5 SPTLC2;ZNF516;RORC
[1] "GO_Cellular_Component_2021"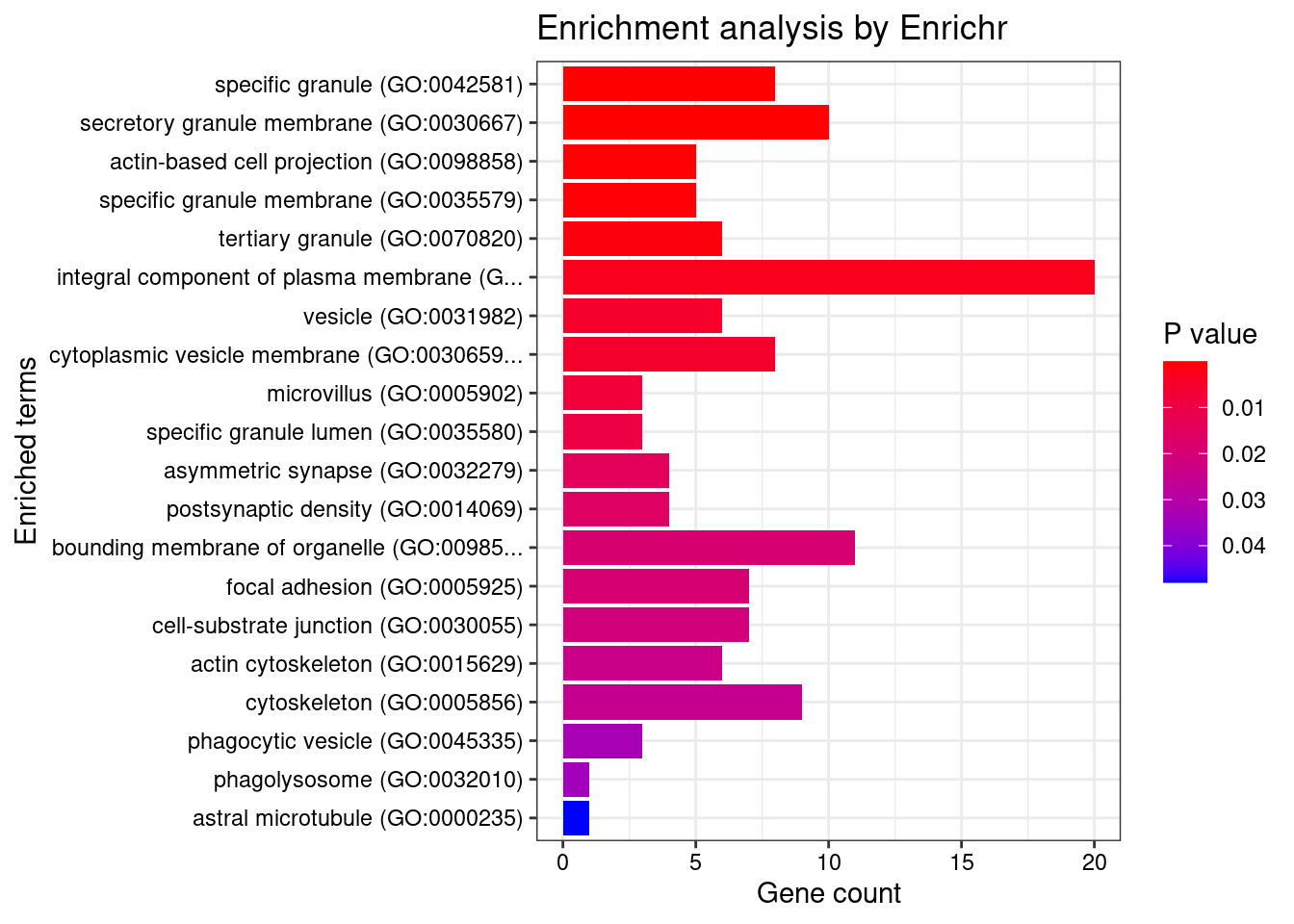
| Version | Author | Date |
|---|---|---|
| 1b50100 | sq-96 | 2023-02-07 |
Term Overlap Adjusted.P.value
1 specific granule (GO:0042581) 8/160 0.001436
2 secretory granule membrane (GO:0030667) 10/274 0.001436
3 actin-based cell projection (GO:0098858) 5/83 0.011822
4 specific granule membrane (GO:0035579) 5/91 0.013581
5 tertiary granule (GO:0070820) 6/164 0.025494
Genes
1 CNN2;ADAM10;PTPRJ;CD36;LYZ;ITGAL;ELANE;CD33
2 PTPRN2;LAMP1;SELL;ADAM10;PTPRJ;CD36;IQGAP2;ITGAL;SIRPB1;CD33
3 SLC9A3R1;MYO1C;CD302;IQGAP2;MYO1G
4 ADAM10;PTPRJ;CD36;ITGAL;CD33
5 CNN2;PTPRN2;LAMP1;ADAM10;LYZ;CD33
[1] "GO_Molecular_Function_2021"
| Version | Author | Date |
|---|---|---|
| 1b50100 | sq-96 | 2023-02-07 |
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)DisGeNET enrichment analysis for genes with PIP>0.5
Description FDR Ratio BgRatio
14 Refractory anaemia with excess blasts 0.09276 1/83 1/9703
45 Cholesterol Ester Storage Disease 0.09276 1/83 1/9703
71 Epistaxis 0.09276 1/83 1/9703
77 Freckles 0.09276 1/83 1/9703
108 Acute Promyelocytic Leukemia 0.09276 3/83 46/9703
111 Liver Cirrhosis, Experimental 0.09276 15/83 774/9703
121 Melanosis 0.09276 1/83 1/9703
122 Chloasma 0.09276 1/83 1/9703
175 Telangiectasis 0.09276 1/83 1/9703
186 Wolman Disease 0.09276 1/83 1/9703WebGestalt enrichment analysis for genes with PIP>0.5
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum =
minNum, : No significant gene set is identified based on FDR 0.05!NULLLoading required package: S4VectorsLoading required package: stats4Loading required package: BiocGenerics
Attaching package: 'BiocGenerics'The following objects are masked from 'package:stats':
IQR, mad, sd, var, xtabsThe following objects are masked from 'package:base':
anyDuplicated, append, as.data.frame, basename, cbind, colnames,
dirname, do.call, duplicated, eval, evalq, Filter, Find, get, grep,
grepl, intersect, is.unsorted, lapply, Map, mapply, match, mget,
order, paste, pmax, pmax.int, pmin, pmin.int, Position, rank,
rbind, Reduce, rownames, sapply, setdiff, sort, table, tapply,
union, unique, unsplit, which.max, which.min
Attaching package: 'S4Vectors'The following objects are masked from 'package:base':
expand.grid, I, unnameLoading required package: IRangesLoading required package: GenomicRangesLoading required package: GenomeInfoDbLoading required package: grida <- locus_plot(region_tag="17_34", return_table=T,
focus=NULL,
label_genes=NULL,
rerun_ctwas=F,
rerun_load_only=F,
label_panel="both",
legend_side="left",
legend_panel="")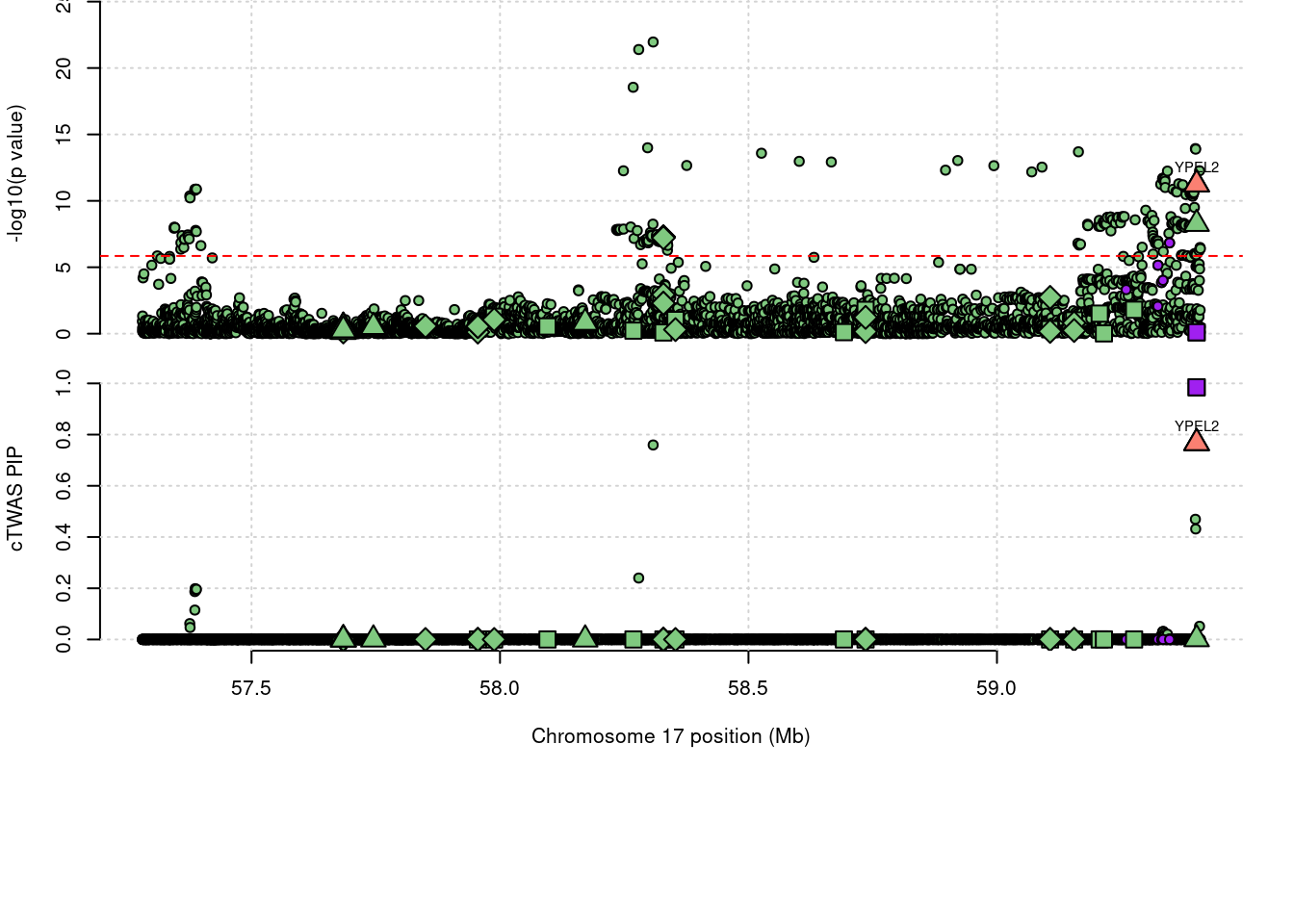
a <- locus_plot(region_tag="19_32", return_table=T,
focus=NULL,
label_genes=NULL,
rerun_ctwas=F,
rerun_load_only=F,
label_panel="both",
legend_side="left",
legend_panel="")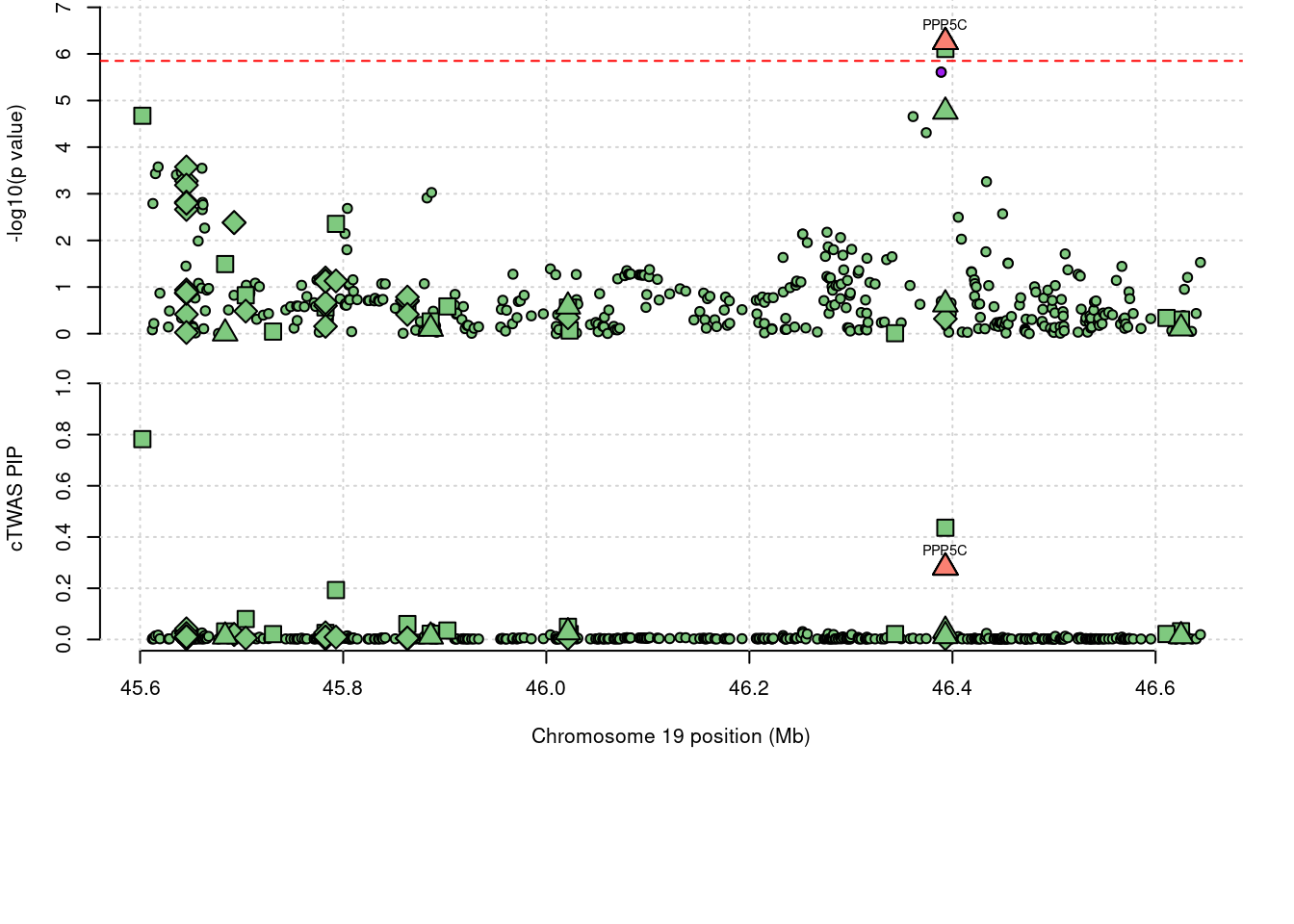
a <- locus_plot(region_tag="12_1", return_table=T,
focus=NULL,
label_genes=NULL,
rerun_ctwas=F,
rerun_load_only=F,
label_panel="both",
legend_side="left",
legend_panel="")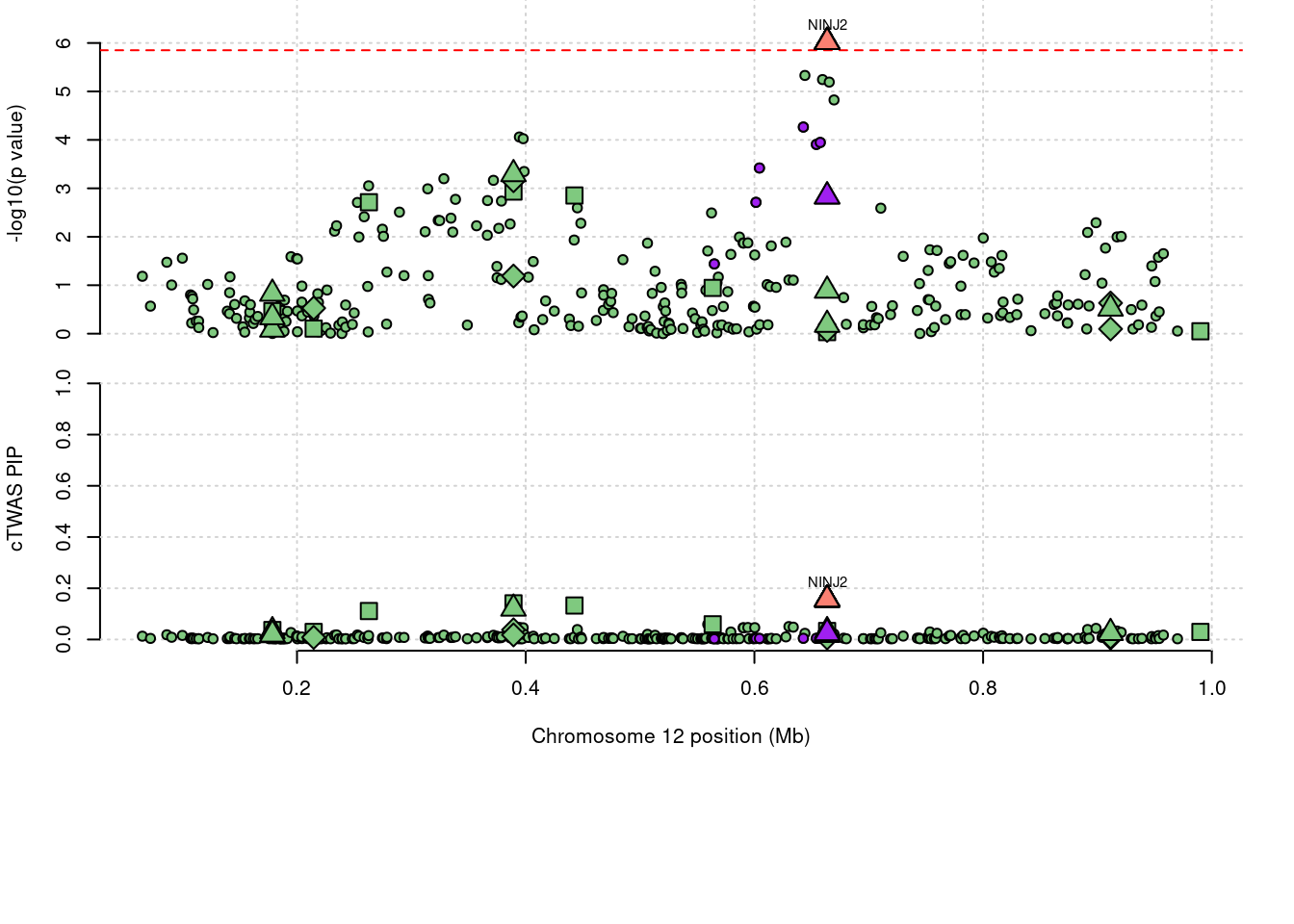
a <- locus_plot(region_tag="1_20", return_table=T,
focus=NULL,
label_genes=NULL,
rerun_ctwas=F,
rerun_load_only=F,
label_panel="both",
legend_side="left",
legend_panel="")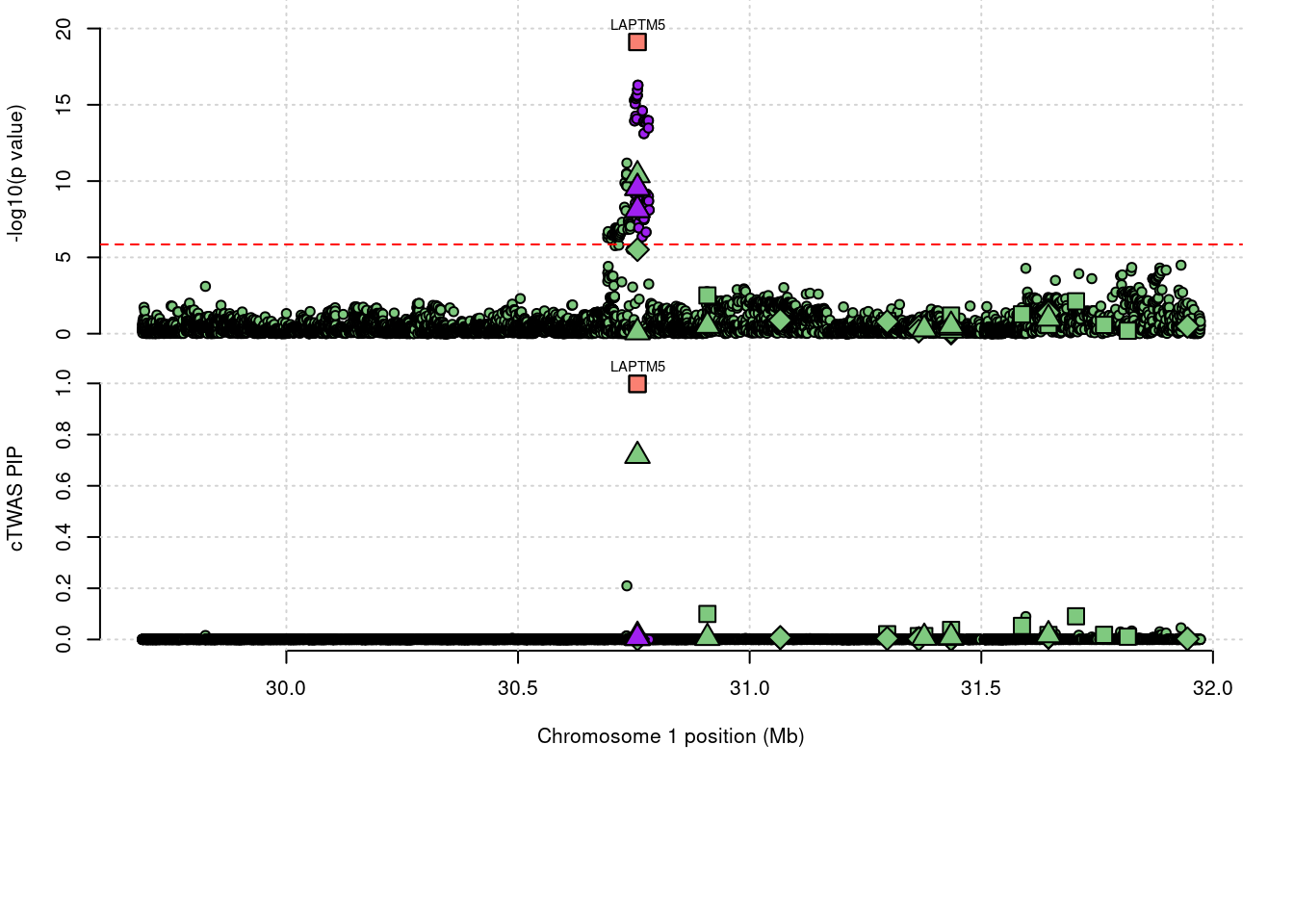
| Version | Author | Date |
|---|---|---|
| 213f0e4 | sq-96 | 2023-02-15 |
sessionInfo()R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] Gviz_1.38.4 GenomicRanges_1.46.1 GenomeInfoDb_1.30.1
[4] IRanges_2.28.0 S4Vectors_0.32.4 BiocGenerics_0.40.0
[7] WebGestaltR_0.4.4 disgenet2r_0.99.2 enrichR_3.1
[10] cowplot_1.1.1 ggplot2_3.4.0 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] backports_1.2.1 Hmisc_4.7-2
[3] BiocFileCache_2.2.1 systemfonts_1.0.4
[5] plyr_1.8.8 igraph_1.3.5
[7] lazyeval_0.2.2 splines_4.1.0
[9] BiocParallel_1.28.3 digest_0.6.31
[11] ensembldb_2.18.4 foreach_1.5.2
[13] htmltools_0.5.4 fansi_1.0.3
[15] checkmate_2.1.0 magrittr_2.0.3
[17] memoise_2.0.1 BSgenome_1.62.0
[19] cluster_2.1.2 doParallel_1.0.17
[21] tzdb_0.3.0 Biostrings_2.62.0
[23] readr_2.1.3 matrixStats_0.63.0
[25] vroom_1.6.0 svglite_2.1.0
[27] prettyunits_1.1.1 jpeg_0.1-10
[29] colorspace_2.0-3 blob_1.2.3
[31] rappdirs_0.3.3 xfun_0.35
[33] dplyr_1.0.10 callr_3.7.3
[35] crayon_1.5.2 RCurl_1.98-1.9
[37] jsonlite_1.8.4 VariantAnnotation_1.40.0
[39] survival_3.2-11 iterators_1.0.14
[41] glue_1.6.2 gtable_0.3.1
[43] zlibbioc_1.40.0 XVector_0.34.0
[45] DelayedArray_0.20.0 apcluster_1.4.10
[47] scales_1.2.1 DBI_1.1.3
[49] rngtools_1.5.2 Rcpp_1.0.9
[51] htmlTable_2.4.1 progress_1.2.2
[53] foreign_0.8-81 bit_4.0.5
[55] Formula_1.2-4 htmlwidgets_1.6.0
[57] httr_1.4.4 RColorBrewer_1.1-3
[59] ellipsis_0.3.2 pkgconfig_2.0.3
[61] XML_3.99-0.13 farver_2.1.0
[63] nnet_7.3-16 sass_0.4.4
[65] dbplyr_2.2.1 deldir_1.0-6
[67] utf8_1.2.2 tidyselect_1.2.0
[69] labeling_0.4.2 rlang_1.0.6
[71] reshape2_1.4.4 later_1.3.0
[73] AnnotationDbi_1.56.2 munsell_0.5.0
[75] tools_4.1.0 cachem_1.0.6
[77] cli_3.4.1 generics_0.1.3
[79] RSQLite_2.2.19 evaluate_0.19
[81] stringr_1.5.0 fastmap_1.1.0
[83] yaml_2.3.6 processx_3.8.0
[85] knitr_1.41 bit64_4.0.5
[87] fs_1.5.2 AnnotationFilter_1.18.0
[89] KEGGREST_1.34.0 doRNG_1.8.2
[91] whisker_0.4.1 xml2_1.3.3
[93] biomaRt_2.50.3 compiler_4.1.0
[95] rstudioapi_0.14 filelock_1.0.2
[97] curl_4.3.2 png_0.1-8
[99] tibble_3.1.8 bslib_0.4.1
[101] stringi_1.7.8 highr_0.9
[103] ps_1.7.2 GenomicFeatures_1.46.5
[105] lattice_0.20-44 ProtGenerics_1.26.0
[107] Matrix_1.3-3 vctrs_0.5.1
[109] pillar_1.8.1 lifecycle_1.0.3
[111] jquerylib_0.1.4 data.table_1.14.6
[113] bitops_1.0-7 httpuv_1.6.7
[115] rtracklayer_1.54.0 R6_2.5.1
[117] BiocIO_1.4.0 latticeExtra_0.6-30
[119] promises_1.2.0.1 gridExtra_2.3
[121] codetools_0.2-18 dichromat_2.0-0.1
[123] assertthat_0.2.1 SummarizedExperiment_1.24.0
[125] rprojroot_2.0.3 rjson_0.2.21
[127] withr_2.5.0 GenomicAlignments_1.30.0
[129] Rsamtools_2.10.0 GenomeInfoDbData_1.2.7
[131] parallel_4.1.0 hms_1.1.2
[133] rpart_4.1-15 rmarkdown_2.19
[135] MatrixGenerics_1.6.0 git2r_0.30.1
[137] biovizBase_1.42.0 getPass_0.2-2
[139] Biobase_2.54.0 base64enc_0.1-3
[141] interp_1.1-3 restfulr_0.0.15