SCZ 2018 - Brain_Anterior_cingulate_cortex_BA24
sheng Qian
2021-2-6
Last updated: 2022-05-19
Checks: 5 2
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version bcaadf3. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .ipynb_checkpoints/
Untracked files:
Untracked: G_list.RData
Untracked: Rplot.png
Untracked: SCZ_annotation.xlsx
Untracked: analysis/.ipynb_checkpoints/
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/Autism_out/
Untracked: code/BMI_S_out/
Untracked: code/BMI_out/
Untracked: code/Glucose_out/
Untracked: code/LDL_S_out/
Untracked: code/SCZ_2014_EUR_out/
Untracked: code/SCZ_2018_S_out/
Untracked: code/SCZ_2018_out/
Untracked: code/SCZ_2020_Single_out/
Untracked: code/SCZ_2020_out/
Untracked: code/SCZ_S_out/
Untracked: code/SCZ_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/process_scz_2018_snps.R
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_Autism_analysis.sbatch
Untracked: code/run_Autism_analysis.sh
Untracked: code/run_Autism_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_analysis_S.sbatch
Untracked: code/run_BMI_analysis_S.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_BMI_ctwas_rss_LDR_S.R
Untracked: code/run_Glucose_analysis.sbatch
Untracked: code/run_Glucose_analysis.sh
Untracked: code/run_Glucose_ctwas_rss_LDR.R
Untracked: code/run_LDL_analysis_S.sbatch
Untracked: code/run_LDL_analysis_S.sh
Untracked: code/run_LDL_ctwas_rss_LDR_S.R
Untracked: code/run_SCZ_2014_EUR_analysis.sbatch
Untracked: code/run_SCZ_2014_EUR_analysis.sh
Untracked: code/run_SCZ_2014_EUR_ctwas_rss_LDR.R
Untracked: code/run_SCZ_2018_analysis.sbatch
Untracked: code/run_SCZ_2018_analysis.sh
Untracked: code/run_SCZ_2018_analysis_S.sbatch
Untracked: code/run_SCZ_2018_analysis_S.sh
Untracked: code/run_SCZ_2018_ctwas_rss_LDR.R
Untracked: code/run_SCZ_2018_ctwas_rss_LDR_S.R
Untracked: code/run_SCZ_2020_Single_analysis.sbatch
Untracked: code/run_SCZ_2020_Single_analysis.sh
Untracked: code/run_SCZ_2020_Single_ctwas_rss_LDR.R
Untracked: code/run_SCZ_2020_analysis.sbatch
Untracked: code/run_SCZ_2020_analysis.sh
Untracked: code/run_SCZ_2020_ctwas_rss_LDR.R
Untracked: code/run_SCZ_analysis.sbatch
Untracked: code/run_SCZ_analysis.sh
Untracked: code/run_SCZ_analysis_S.sbatch
Untracked: code/run_SCZ_analysis_S.sh
Untracked: code/run_SCZ_ctwas_rss_LDR.R
Untracked: code/run_SCZ_ctwas_rss_LDR_S.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: code/wflow_build.R
Untracked: code/wflow_build.sbatch
Untracked: data/.ipynb_checkpoints/
Untracked: data/GO_Terms/
Untracked: data/PGC3_SCZ_wave3_public.v2.tsv
Untracked: data/SCZ/
Untracked: data/SCZ_2014_EUR/
Untracked: data/SCZ_2018/
Untracked: data/SCZ_2018_S/
Untracked: data/SCZ_2020/
Untracked: data/SCZ_S/
Untracked: data/Supplementary Table 15 - MAGMA.xlsx
Untracked: data/Supplementary Table 20 - Prioritised Genes.xlsx
Untracked: data/T2D/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/scz_2018.RDS
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Untracked: top_genes_32.txt
Untracked: top_genes_37.txt
Untracked: top_genes_43.txt
Untracked: top_genes_54.txt
Untracked: top_genes_81.txt
Untracked: z_snp_pos_SCZ.RData
Untracked: z_snp_pos_SCZ_2014_EUR.RData
Untracked: z_snp_pos_SCZ_2018.RData
Untracked: z_snp_pos_SCZ_2020.RData
Unstaged changes:
Deleted: analysis/BMI_S_results.Rmd
Modified: analysis/SCZ_2018_Brain_Amygdala_S.Rmd
Modified: analysis/SCZ_2018_Brain_Anterior_cingulate_cortex_BA24_S.Rmd
Modified: analysis/SCZ_2018_Brain_Caudate_basal_ganglia_S.Rmd
Modified: analysis/SCZ_2018_Brain_Cerebellar_Hemisphere_S.Rmd
Modified: analysis/SCZ_2018_Brain_Cerebellum_S.Rmd
Modified: analysis/SCZ_2018_Brain_Cortex_S.Rmd
Modified: analysis/SCZ_2018_Brain_Frontal_Cortex_BA9_S.Rmd
Modified: analysis/SCZ_2018_Brain_Hippocampus_S.Rmd
Modified: analysis/SCZ_2018_Brain_Hypothalamus_S.Rmd
Modified: analysis/SCZ_2018_Brain_Nucleus_accumbens_basal_ganglia_S.Rmd
Modified: analysis/SCZ_2018_Brain_Putamen_basal_ganglia_S.Rmd
Modified: analysis/SCZ_2018_Brain_Spinal_cord_cervical_c-1_S.Rmd
Modified: analysis/SCZ_2018_Brain_Substantia_nigra_S.Rmd
Modified: analysis/ttt.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/SCZ_2018_Brain_Anterior_cingulate_cortex_BA24_S.Rmd) and HTML (docs/SCZ_2018_Brain_Anterior_cingulate_cortex_BA24_S.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | bcaadf3 | sq-96 | 2022-05-19 | update |
| html | bcaadf3 | sq-96 | 2022-05-19 | update |
| Rmd | be614ed | sq-96 | 2022-05-19 | update |
| html | be614ed | sq-96 | 2022-05-19 | update |
| Rmd | 7d08c9b | sq-96 | 2022-05-18 | update |
| html | 7d08c9b | sq-96 | 2022-05-18 | update |
| Rmd | 2749be9 | sq-96 | 2022-05-12 | update |
| html | 2749be9 | sq-96 | 2022-05-12 | update |
| html | 011327d | sq-96 | 2022-05-12 | update |
| Rmd | 6c6abbd | sq-96 | 2022-05-12 | update |
library(reticulate)
use_python("/scratch/midway2/shengqian/miniconda3/envs/PythonForR/bin/python",required=T)Weight QC
#number of imputed weights
nrow(qclist_all)[1] 18988#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
1766 1297 1118 729 754 987 1093 674 787 871 1192 1048 389 666 643 793
17 18 19 20 21 22
1329 261 1342 620 35 594 #number of imputed weights without missing variants
sum(qclist_all$nmiss==0)[1] 16837#proportion of imputed weights without missing variants
mean(qclist_all$nmiss==0)[1] 0.8867INFO:numexpr.utils:Note: NumExpr detected 56 cores but "NUMEXPR_MAX_THREADS" not set, so enforcing safe limit of 8.finish
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
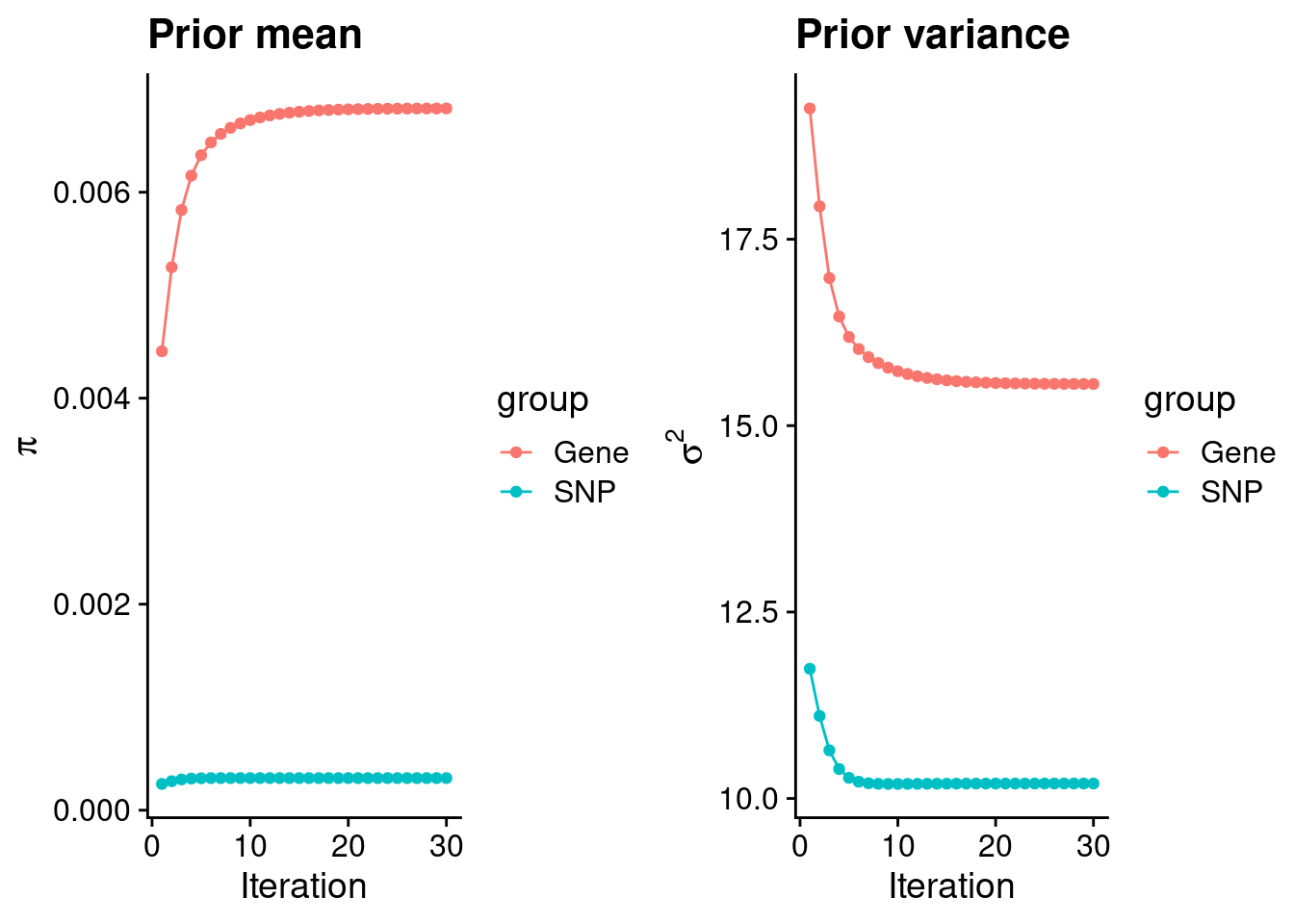
intersect, setdiff, setequal, unionCheck convergence of parameters
| Version | Author | Date |
|---|---|---|
| 2749be9 | sq-96 | 2022-05-12 |
gene snp
0.0068134 0.0003114 gene snp
15.56 10.20 [1] 105318[1] 7115 6309950 gene snp
0.007161 0.190316 [1] 0.01572 1.10677Genes with highest PIPs
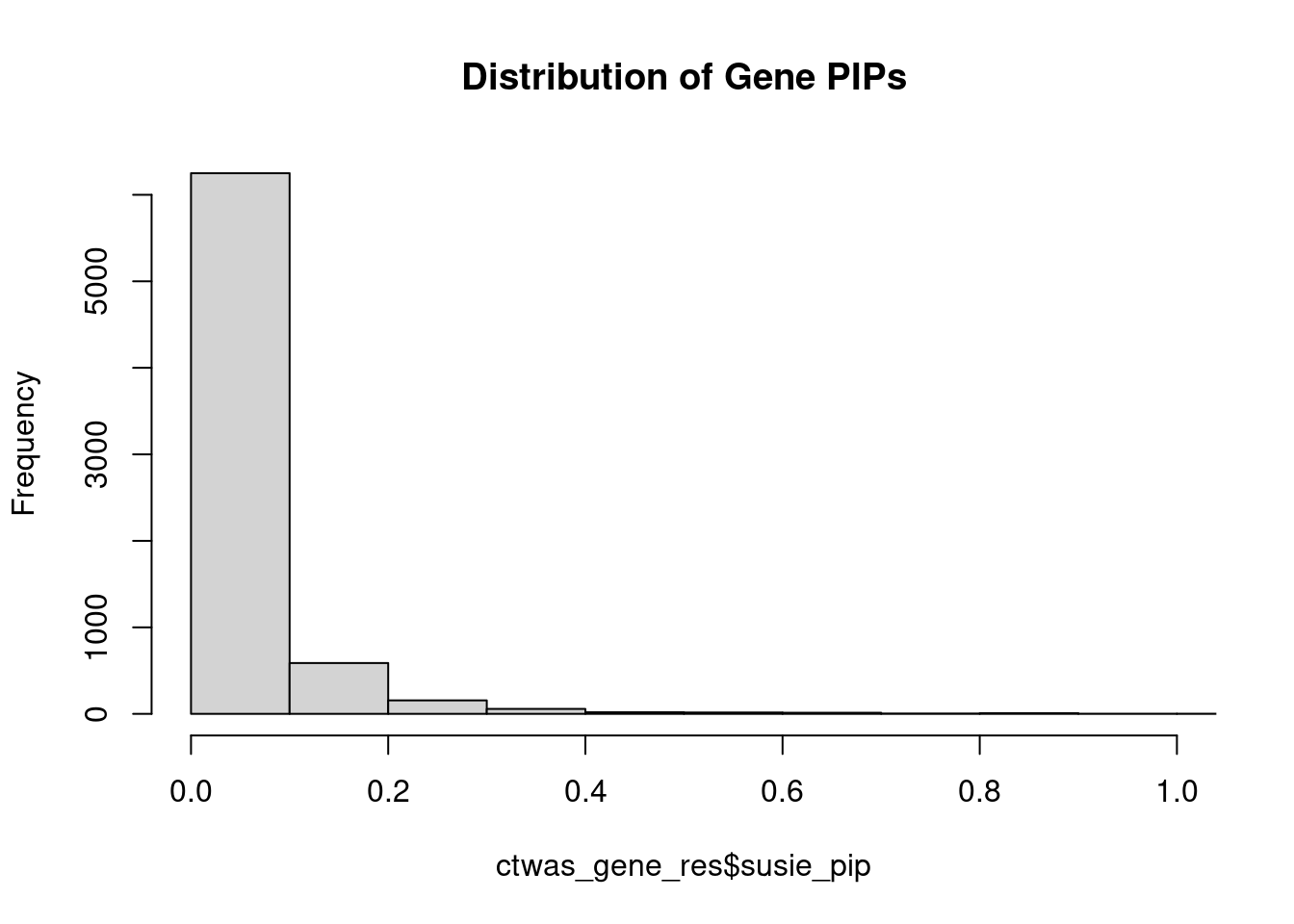
genename region_tag susie_pip mu2 PVE z num_intron
765 BTN2A1 6_20 1.0491 157.37 1.570e-03 -13.238 6
3347 LRP8 1_33 1.0010 25.97 2.331e-04 4.820 6
3331 LPCAT4 15_10 0.9457 26.08 2.141e-04 4.892 4
772 BUB1B-PAK6 15_14 0.9157 31.13 2.438e-04 -5.588 3
4084 NRXN2 11_36 0.8999 25.63 1.950e-04 4.723 4
5808 SPECC1 17_16 0.8947 26.90 2.013e-04 -5.038 3
627 B3GAT1 11_84 0.8865 22.73 1.578e-04 4.343 9
3740 MRPS33 7_87 0.8542 21.83 1.489e-04 -4.304 4
6155 THAP8 19_25 0.8477 20.15 1.375e-04 3.847 2
265 AKT3 1_128 0.8362 36.35 2.280e-04 -6.350 7
419 APOPT1 14_54 0.8209 50.53 3.169e-04 7.429 7
691 BDNF 11_19 0.8143 24.21 1.499e-04 4.348 3
1867 DPYSL3 5_86 0.7890 23.30 1.377e-04 4.157 1
2489 GIGYF1 7_62 0.7603 28.78 1.561e-04 5.266 2
3985 NGEF 2_137 0.7358 31.04 1.576e-04 6.994 2
1656 DBF4B 17_26 0.7211 19.92 9.437e-05 -3.890 5
4832 R3HDM2 12_36 0.6939 22.64 1.027e-04 -4.237 2
5749 SNRPA1 15_50 0.6925 22.95 1.029e-04 -3.948 2
2820 HSPA9 5_82 0.6773 28.81 1.255e-04 5.633 1
138 ACTR1B 2_57 0.6669 21.84 9.034e-05 -3.978 4
num_sqtl
765 7
3347 6
3331 4
772 3
4084 4
5808 4
627 14
3740 4
6155 2
265 8
419 9
691 4
1867 1
2489 2
3985 2
1656 5
4832 3
5749 3
2820 1
138 4Genes with highest PVE
genename region_tag susie_pip mu2 PVE z num_intron
765 BTN2A1 6_20 1.0491 157.37 0.0015695 -13.238 6
3381 LSM2 6_26 0.3709 658.01 0.0008595 -11.599 1
418 APOM 6_26 0.2474 649.69 0.0003774 11.590 2
419 APOPT1 14_54 0.8209 50.53 0.0003169 7.429 7
772 BUB1B-PAK6 15_14 0.9157 31.13 0.0002438 -5.588 3
3347 LRP8 1_33 1.0010 25.97 0.0002331 4.820 6
265 AKT3 1_128 0.8362 36.35 0.0002280 -6.350 7
3331 LPCAT4 15_10 0.9457 26.08 0.0002141 4.892 4
5808 SPECC1 17_16 0.8947 26.90 0.0002013 -5.038 3
4084 NRXN2 11_36 0.8999 25.63 0.0001950 4.723 4
627 B3GAT1 11_84 0.8865 22.73 0.0001578 4.343 9
3985 NGEF 2_137 0.7358 31.04 0.0001576 6.994 2
2489 GIGYF1 7_62 0.7603 28.78 0.0001561 5.266 2
3751 MSH5 6_26 0.1568 654.60 0.0001528 11.531 2
691 BDNF 11_19 0.8143 24.21 0.0001499 4.348 3
3740 MRPS33 7_87 0.8542 21.83 0.0001489 -4.304 4
1867 DPYSL3 5_86 0.7890 23.30 0.0001377 4.157 1
6155 THAP8 19_25 0.8477 20.15 0.0001375 3.847 2
6685 VARS 6_26 0.1464 650.58 0.0001324 -11.548 1
2308 FES 15_42 0.6005 37.84 0.0001270 5.964 3
num_sqtl
765 7
3381 1
418 2
419 9
772 3
3347 6
265 8
3331 4
5808 4
4084 4
627 14
3985 2
2489 2
3751 2
691 4
3740 4
1867 1
6155 2
6685 1
2308 4Comparing z scores and PIPs
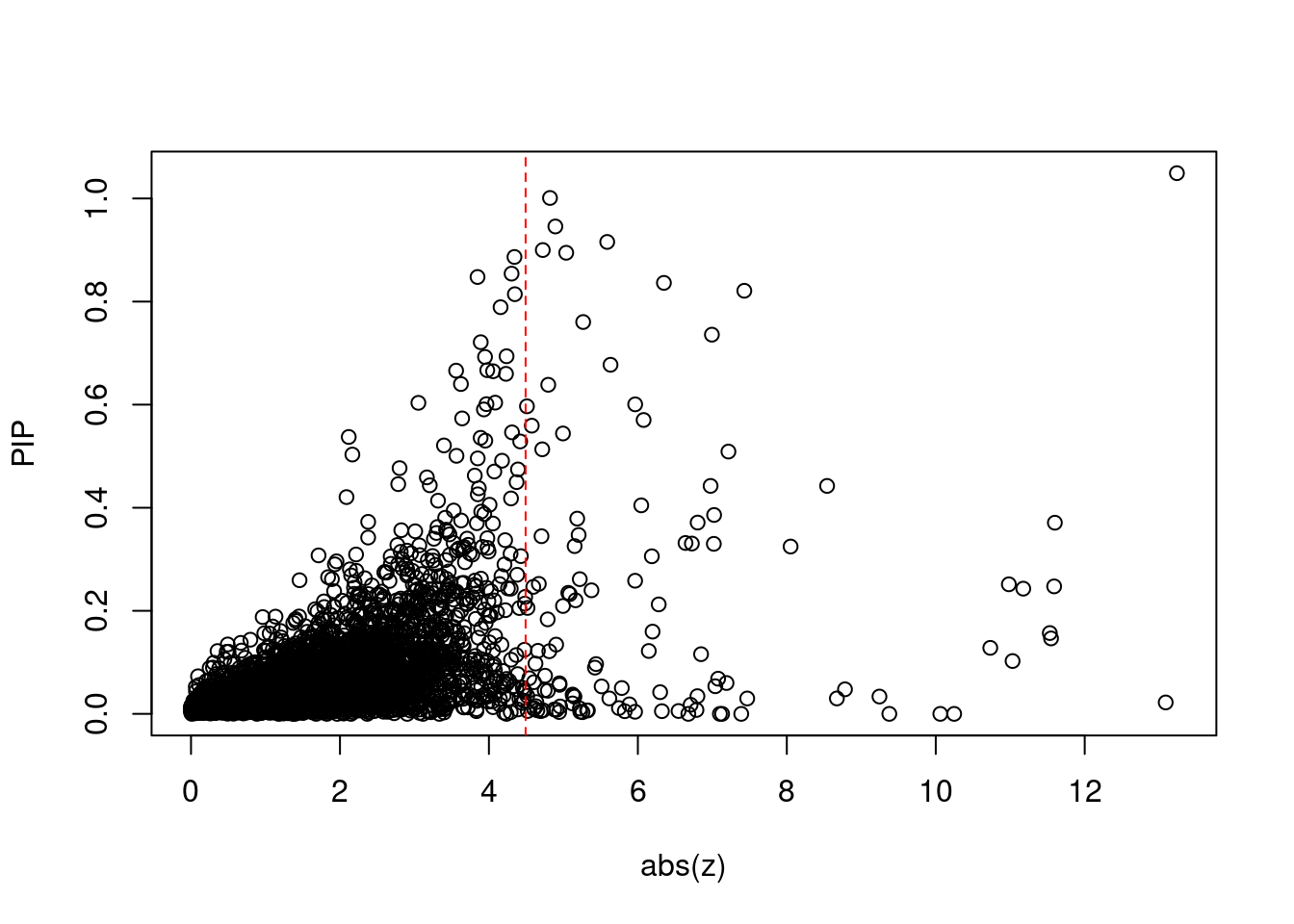
[1] 0.01673 genename region_tag susie_pip mu2 PVE z num_intron
765 BTN2A1 6_20 1.049e+00 157.37 1.570e-03 -13.238 6
4373 PGBD1 6_22 2.219e-02 167.48 4.926e-07 -13.087 2
3381 LSM2 6_26 3.709e-01 658.01 8.595e-04 -11.599 1
418 APOM 6_26 2.474e-01 649.69 3.774e-04 11.590 2
6685 VARS 6_26 1.464e-01 650.58 1.324e-04 -11.548 1
3751 MSH5 6_26 1.568e-01 654.60 1.528e-04 11.531 2
1693 DDR1 6_25 2.429e-01 105.31 5.771e-05 11.175 3
894 C6orf136 6_24 1.025e-01 84.83 8.464e-06 11.031 2
2345 FLOT1 6_24 2.514e-01 83.46 4.991e-05 10.981 7
768 BTN3A2 6_20 1.281e-01 102.85 5.057e-06 -10.732 5
643 BAG6 6_26 8.739e-10 518.30 3.759e-21 10.247 7
4649 PPT2 6_26 4.838e-13 483.22 1.074e-27 -10.061 4
2597 GPSM3 6_26 2.154e-14 431.49 1.901e-30 9.377 2
1084 CCHCR1 6_25 3.360e-02 63.97 3.100e-07 -9.244 9
2755 HLA-DMA 6_27 4.764e-02 70.44 8.914e-07 8.781 4
5239 RP5-874C20.8 6_22 2.995e-02 56.35 2.939e-07 8.672 4
4099 NT5C2 10_66 4.423e-01 49.30 8.938e-05 -8.541 9
513 AS3MT 10_66 3.247e-01 46.06 4.535e-05 8.051 5
7107 ZSCAN16 6_22 2.986e-02 56.18 3.388e-07 7.468 4
419 APOPT1 14_54 8.209e-01 50.53 3.169e-04 7.429 7
num_sqtl
765 7
4373 2
3381 1
418 2
6685 1
3751 2
1693 3
894 2
2345 7
768 6
643 7
4649 4
2597 2
1084 13
2755 4
5239 4
4099 12
513 5
7107 4
419 9GO enrichment analysis for genes with PIP>0.5
#number of genes for gene set enrichment
length(genes)[1] 45Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"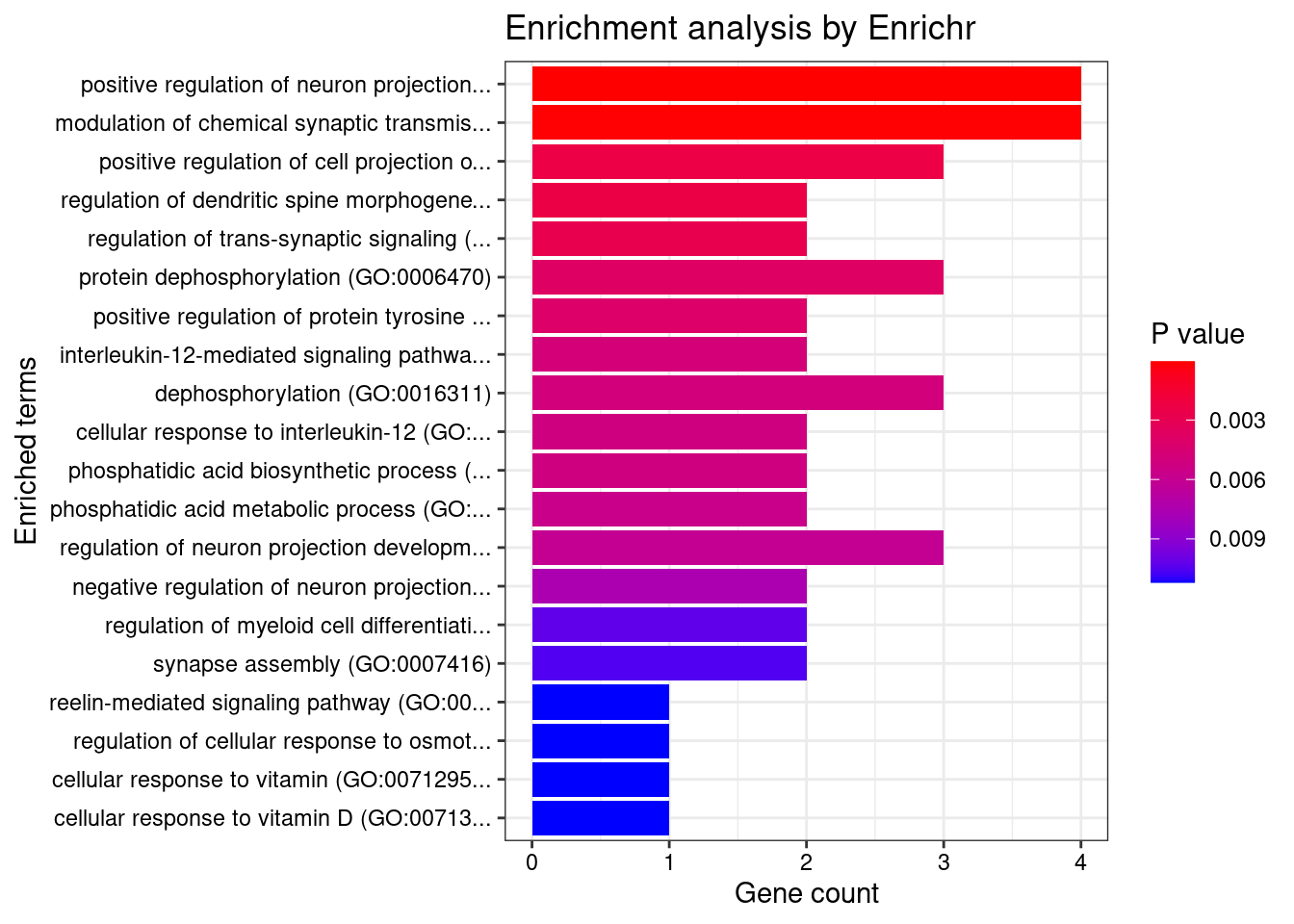
Term Overlap
1 positive regulation of neuron projection development (GO:0010976) 4/88
2 modulation of chemical synaptic transmission (GO:0050804) 4/109
Adjusted.P.value Genes
1 0.01853 BDNF;FES;DPYSL3;LRP8
2 0.02136 BDNF;LRP8;DGKZ;BEGAIN
[1] "GO_Cellular_Component_2021"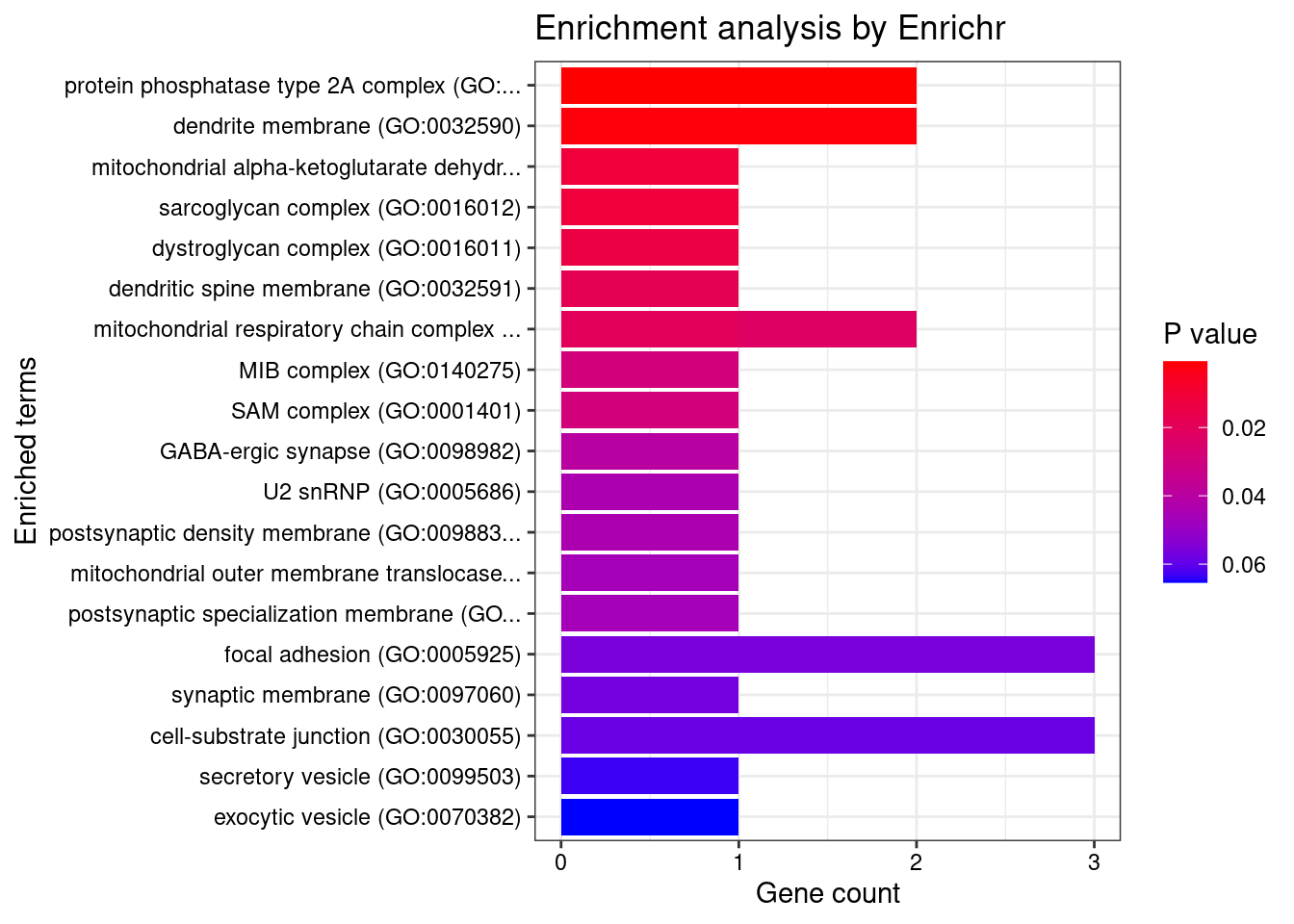
Term Overlap Adjusted.P.value
1 protein phosphatase type 2A complex (GO:0000159) 2/17 0.03624
Genes
1 PPP2R5B;PPP2R2A
[1] "GO_Molecular_Function_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)DisGeNET enrichment analysis for genes with PIP>0.5
Description FDR
12 Renal Cell Carcinoma 0.0342
36 Measles 0.0342
137 Chromophobe Renal Cell Carcinoma 0.0342
138 Sarcomatoid Renal Cell Carcinoma 0.0342
139 Collecting Duct Carcinoma of the Kidney 0.0342
142 Papillary Renal Cell Carcinoma 0.0342
152 Maple Syrup Urine Disease, Type IA 0.0342
157 HEMOLYTIC UREMIC SYNDROME, ATYPICAL, SUSCEPTIBILITY TO, 2 0.0342
166 MITOCHONDRIAL COMPLEX III DEFICIENCY, NUCLEAR TYPE 5 0.0342
174 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 2 0.0342
Ratio BgRatio
12 3/20 128/9703
36 1/20 1/9703
137 3/20 128/9703
138 3/20 128/9703
139 3/20 128/9703
142 3/20 128/9703
152 1/20 1/9703
157 1/20 1/9703
166 1/20 1/9703
174 1/20 1/9703WebGestalt enrichment analysis for genes with PIP>0.5
Warning: replacing previous import 'lifecycle::last_warnings' by
'rlang::last_warnings' when loading 'hms'Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum =
minNum, : No significant gene set is identified based on FDR 0.05!NULLPIP Manhattan Plot
Warning: ggrepel: 5 unlabeled data points (too many overlaps). Consider
increasing max.overlaps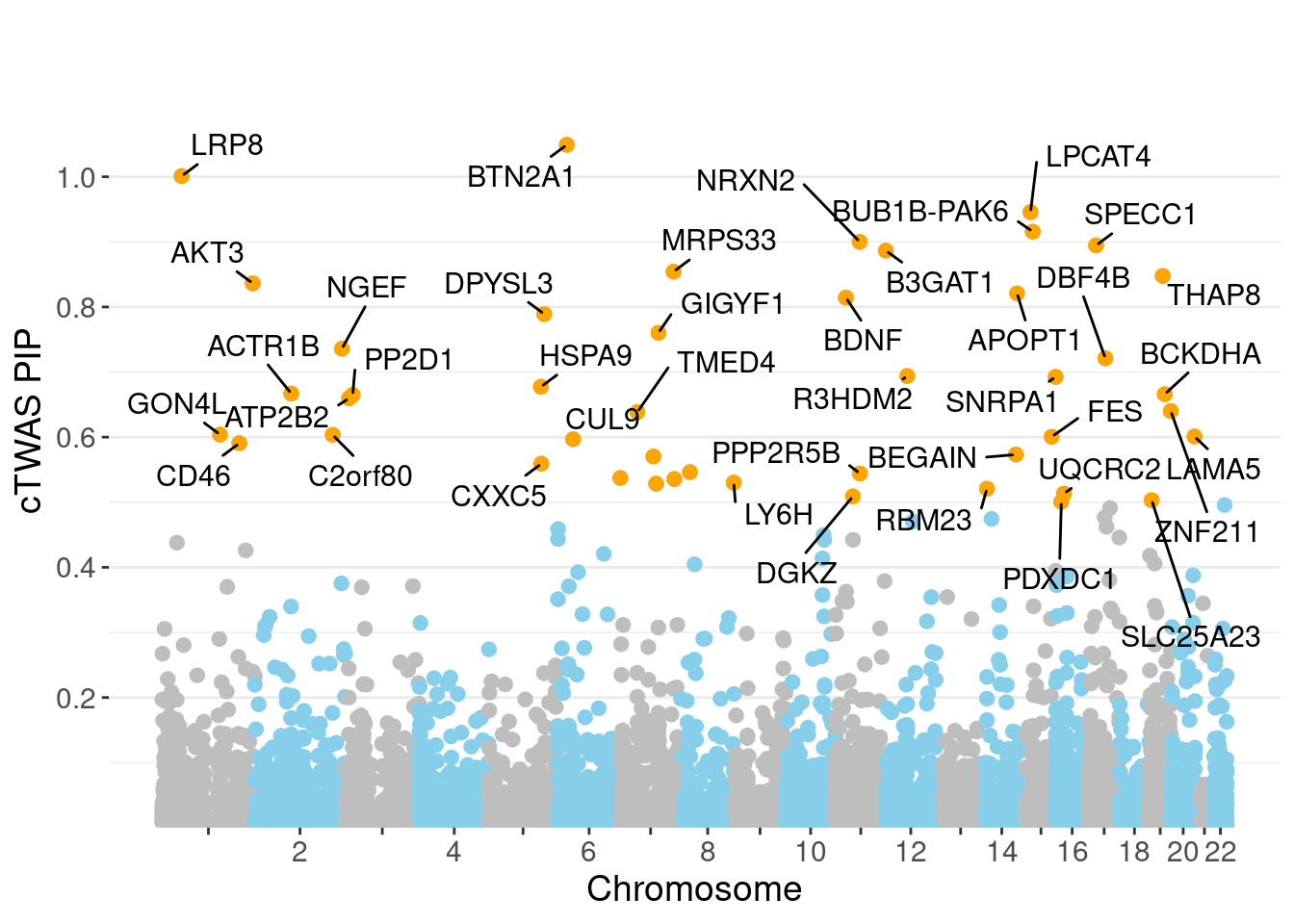
Sensitivity, specificity and precision for silver standard genes
#number of genes in known annotations
print(length(known_annotations))[1] 130#number of genes in known annotations with imputed expression
print(sum(known_annotations %in% ctwas_gene_res$genename))[1] 51#significance threshold for TWAS
print(sig_thresh)[1] 4.493#number of ctwas genes
length(ctwas_genes)[1] 12#number of TWAS genes
length(twas_genes)[1] 119#show novel genes (ctwas genes with not in TWAS genes)
ctwas_gene_res[ctwas_gene_res$genename %in% novel_genes,report_cols] genename region_tag susie_pip mu2 PVE z num_intron num_sqtl
627 B3GAT1 11_84 0.8865 22.73 0.0001578 4.343 9 14
691 BDNF 11_19 0.8143 24.21 0.0001499 4.348 3 4
3740 MRPS33 7_87 0.8542 21.83 0.0001489 -4.304 4 4
6155 THAP8 19_25 0.8477 20.15 0.0001375 3.847 2 2#sensitivity / recall
print(sensitivity) ctwas TWAS
0.01538 0.08462 #specificity
print(specificity) ctwas TWAS
0.9986 0.9847 #precision / PPV
print(precision) ctwas TWAS
0.16667 0.09244
sessionInfo()R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] readxl_1.4.0 forcats_0.5.1 stringr_1.4.0 purrr_0.3.4
[5] readr_1.4.0 tidyr_1.1.3 tidyverse_1.3.1 tibble_3.1.7
[9] WebGestaltR_0.4.4 disgenet2r_0.99.2 enrichR_3.0 cowplot_1.1.1
[13] ggplot2_3.3.5 dplyr_1.0.7 reticulate_1.25 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] fs_1.5.0 lubridate_1.7.10 doParallel_1.0.16 httr_1.4.2
[5] rprojroot_2.0.2 tools_4.1.0 backports_1.2.1 doRNG_1.8.2
[9] bslib_0.2.5.1 utf8_1.2.1 R6_2.5.0 vipor_0.4.5
[13] DBI_1.1.1 colorspace_2.0-2 withr_2.4.2 ggrastr_1.0.1
[17] tidyselect_1.1.1 processx_3.5.2 curl_4.3.2 compiler_4.1.0
[21] git2r_0.28.0 rvest_1.0.0 cli_3.0.0 Cairo_1.5-15
[25] xml2_1.3.2 labeling_0.4.2 sass_0.4.0 scales_1.1.1
[29] callr_3.7.0 systemfonts_1.0.4 apcluster_1.4.9 digest_0.6.27
[33] rmarkdown_2.9 svglite_2.0.0 pkgconfig_2.0.3 htmltools_0.5.1.1
[37] dbplyr_2.1.1 highr_0.9 rlang_1.0.2 rstudioapi_0.13
[41] jquerylib_0.1.4 farver_2.1.0 generics_0.1.0 jsonlite_1.7.2
[45] magrittr_2.0.1 Matrix_1.3-3 ggbeeswarm_0.6.0 Rcpp_1.0.7
[49] munsell_0.5.0 fansi_0.5.0 lifecycle_1.0.0 stringi_1.6.2
[53] whisker_0.4 yaml_2.2.1 plyr_1.8.6 grid_4.1.0
[57] ggrepel_0.9.1 parallel_4.1.0 promises_1.2.0.1 crayon_1.4.1
[61] lattice_0.20-44 haven_2.4.1 hms_1.1.0 knitr_1.33
[65] ps_1.6.0 pillar_1.7.0 igraph_1.2.6 rjson_0.2.20
[69] rngtools_1.5 reshape2_1.4.4 codetools_0.2-18 reprex_2.0.0
[73] glue_1.4.2 evaluate_0.14 getPass_0.2-2 modelr_0.1.8
[77] data.table_1.14.0 png_0.1-7 vctrs_0.3.8 httpuv_1.6.1
[81] foreach_1.5.1 cellranger_1.1.0 gtable_0.3.0 assertthat_0.2.1
[85] xfun_0.24 broom_0.7.8 later_1.2.0 iterators_1.0.13
[89] beeswarm_0.4.0 ellipsis_0.3.2 here_1.0.1