LDL - Liver Traits Jointly
Sheng Qian
2023-2-1
Last updated: 2023-06-02
Checks: 5 2
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config_b38.R | code/ctwas_config_b38.R |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/Predictive_Models/Liver_E.db | data/Predictive_Models/Liver_E.db |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/Predictive_Models/Liver_Splicing_mapping.RData | data/Predictive_Models/Liver_Splicing_mapping.RData |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/Predictive_Models/WholeBlood_M.db | data/Predictive_Models/WholeBlood_M.db |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/G_list.RData | data/G_list.RData |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/locus_plot.R | code/locus_plot.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 3d0d6e3. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .ipynb_checkpoints/
Ignored: analysis/figure/
Untracked files:
Untracked: LDL_LDLR_genetrack.pdf
Untracked: LDL_LDLR_locus.pdf
Untracked: LDL_TEME199_genetrack.pdf
Untracked: LDL_TEME199_locus.pdf
Untracked: Proposal plots.R
Untracked: RGS14.pdf
Untracked: RNF186.pdf
Untracked: Rplots.pdf
Untracked: SCZ_annotation.xlsx
Untracked: SLC8B1.pdf
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/LDL_Liver_E_S_M.Rmd
Untracked: analysis/LungCancer_Lung_E_S_M.Rmd
Untracked: analysis/ProstateCancer_Prostate_E_S_M.Rmd
Untracked: cache/
Untracked: code/.ipynb_checkpoints/
Untracked: code/LDL_E_S_M_out/
Untracked: code/LungCancer_E_S_M_out/
Untracked: code/ProstateCancer_E_S_M_out/
Untracked: code/WhiteBlood_E_S_M_PC_out/
Untracked: code/run_LDL_analysis_E_S_M.sbatch
Untracked: code/run_LDL_analysis_E_S_M.sh
Untracked: code/run_LDL_ctwas_rss_LDR_E_S_M.R
Untracked: code/run_LungCancer_analysis_E_S_M.sbatch
Untracked: code/run_LungCancer_analysis_E_S_M.sh
Untracked: code/run_LungCancer_ctwas_rss_LDR_E_S_M.R
Untracked: code/run_ProstateCancer_analysis_E_S_M.sbatch
Untracked: code/run_ProstateCancer_analysis_E_S_M.sh
Untracked: code/run_ProstateCancer_ctwas_rss_LDR_E_S_M.R
Untracked: data/.ipynb_checkpoints/
Untracked: data/FUMA_output/
Untracked: data/GO_Terms/
Untracked: data/GTEx_Analysis_v8_eQTL.tar
Untracked: data/G_list.RData
Untracked: data/IBD_ME/
Untracked: data/LDL/
Untracked: data/LDL_E_S/
Untracked: data/LDL_E_S_M/
Untracked: data/LDL_M/
Untracked: data/LDL_S/
Untracked: data/LungCancer_E_S_M/
Untracked: data/PGC3_SCZ_wave3_public.v2.tsv
Untracked: data/Predictive_Models/
Untracked: data/ProstateCancer_E_S_M/
Untracked: data/Supplementary Table 15 - MAGMA.xlsx
Untracked: data/Supplementary Table 20 - Prioritised Genes.xlsx
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/WhiteBlood_E/
Untracked: data/WhiteBlood_E_M/
Untracked: data/WhiteBlood_E_S_M/
Untracked: data/WhiteBlood_E_S_M_PC/
Untracked: data/WhiteBlood_M/
Untracked: data/WhiteBlood_M_compare/
Untracked: data/WhiteBlood_M_enet/
Untracked: data/WhiteBlood_M_mashr/
Untracked: data/cpg_annot.RData
Untracked: data/eqtl/
Untracked: data/gencode.v26.GRCh38.genes.gtf
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/gwas_sumstats/
Untracked: data/magma.genes.out
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/mqtl/
Untracked: data/notes.txt
Untracked: data/scz_2018.RDS
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/test/
Untracked: hist.pdf
Untracked: submit.sh
Untracked: temp.regionlist.RDS
Untracked: temp.regions.txt
Untracked: temp.susieIrss.txt
Untracked: temp.temp.susieIrssres.Rd
Untracked: temp_LDR/
Untracked: temp_ld_R_chr1.txt
Untracked: temp_ld_R_chr10.txt
Untracked: temp_ld_R_chr11.txt
Untracked: temp_ld_R_chr12.txt
Untracked: temp_ld_R_chr13.txt
Untracked: temp_ld_R_chr14.txt
Untracked: temp_ld_R_chr15.txt
Untracked: temp_ld_R_chr16.txt
Untracked: temp_ld_R_chr17.txt
Untracked: temp_ld_R_chr18.txt
Untracked: temp_ld_R_chr19.txt
Untracked: temp_ld_R_chr2.txt
Untracked: temp_ld_R_chr20.txt
Untracked: temp_ld_R_chr21.txt
Untracked: temp_ld_R_chr22.txt
Untracked: temp_ld_R_chr3.txt
Untracked: temp_ld_R_chr4.txt
Untracked: temp_ld_R_chr5.txt
Untracked: temp_ld_R_chr6.txt
Untracked: temp_ld_R_chr7.txt
Untracked: temp_ld_R_chr8.txt
Untracked: temp_ld_R_chr9.txt
Untracked: temp_reg.txt
Unstaged changes:
Deleted: analysis/Atrial_Fibrillation_Heart_Atrial_Appendage.Rmd
Deleted: analysis/Atrial_Fibrillation_Heart_Left_Ventricle.Rmd
Deleted: analysis/Autism_Brain_Amygdala.Rmd
Deleted: analysis/Autism_Brain_Anterior_cingulate_cortex_BA24.Rmd
Deleted: analysis/Autism_Brain_Caudate_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Cerebellar_Hemisphere.Rmd
Deleted: analysis/Autism_Brain_Cerebellum.Rmd
Deleted: analysis/Autism_Brain_Cortex.Rmd
Deleted: analysis/Autism_Brain_Frontal_Cortex_BA9.Rmd
Deleted: analysis/Autism_Brain_Hippocampus.Rmd
Deleted: analysis/Autism_Brain_Hypothalamus.Rmd
Deleted: analysis/Autism_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Putamen_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Spinal_cord_cervical_c-1.Rmd
Deleted: analysis/Autism_Brain_Substantia_nigra.Rmd
Deleted: analysis/BMI_Brain_Amygdala.Rmd
Deleted: analysis/BMI_Brain_Amygdala_S.Rmd
Deleted: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24.Rmd
Deleted: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24_S.Rmd
Deleted: analysis/BMI_Brain_Caudate_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Caudate_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Cerebellar_Hemisphere.Rmd
Deleted: analysis/BMI_Brain_Cerebellar_Hemisphere_S.Rmd
Deleted: analysis/BMI_Brain_Cerebellum.Rmd
Deleted: analysis/BMI_Brain_Cerebellum_S.Rmd
Deleted: analysis/BMI_Brain_Cortex.Rmd
Deleted: analysis/BMI_Brain_Cortex_S.Rmd
Deleted: analysis/BMI_Brain_Frontal_Cortex_BA9.Rmd
Deleted: analysis/BMI_Brain_Frontal_Cortex_BA9_S.Rmd
Deleted: analysis/BMI_Brain_Hippocampus.Rmd
Deleted: analysis/BMI_Brain_Hippocampus_S.Rmd
Deleted: analysis/BMI_Brain_Hypothalamus.Rmd
Deleted: analysis/BMI_Brain_Hypothalamus_S.Rmd
Deleted: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Putamen_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Putamen_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Spinal_cord_cervical_c-1.Rmd
Deleted: analysis/BMI_Brain_Spinal_cord_cervical_c-1_S.Rmd
Deleted: analysis/BMI_Brain_Substantia_nigra.Rmd
Deleted: analysis/BMI_Brain_Substantia_nigra_S.Rmd
Deleted: analysis/BMI_S_results.Rmd
Deleted: analysis/Glucose_Adipose_Subcutaneous.Rmd
Deleted: analysis/Glucose_Adipose_Visceral_Omentum.Rmd
Modified: analysis/LD_block_analysis.Rmd
Modified: analysis/LD_block_analysis_merge1000bp.Rmd
Modified: analysis/index.Rmd
Deleted: code/WhiteBlood_M_ener_out/WhiteBlood_WholeBlood.out
Deleted: code/White_Blood_M_out/White_Blood_BreastMammary.err
Deleted: code/White_Blood_M_out/White_Blood_BreastMammary.out
Deleted: code/White_Blood_M_out/White_Blood_ColonTransverse.err
Deleted: code/White_Blood_M_out/White_Blood_ColonTransverse.out
Deleted: code/White_Blood_M_out/White_Blood_KidneyCortex.err
Deleted: code/White_Blood_M_out/White_Blood_KidneyCortex.out
Deleted: code/White_Blood_M_out/White_Blood_Lung.err
Deleted: code/White_Blood_M_out/White_Blood_Lung.out
Deleted: code/White_Blood_M_out/White_Blood_MuscleSkeletal.err
Deleted: code/White_Blood_M_out/White_Blood_MuscleSkeletal.out
Deleted: code/White_Blood_M_out/White_Blood_Ovary.err
Deleted: code/White_Blood_M_out/White_Blood_Ovary.out
Deleted: code/White_Blood_M_out/White_Blood_Prostate.err
Deleted: code/White_Blood_M_out/White_Blood_Prostate.out
Deleted: code/White_Blood_M_out/White_Blood_Testis.err
Deleted: code/White_Blood_M_out/White_Blood_Testis.out
Deleted: code/White_Blood_M_out/White_Blood_WholeBlood.err
Deleted: code/White_Blood_M_out/White_Blood_WholeBlood.out
Modified: code/ctwas_config_b38.R
Deleted: code/run_IBD_ctwas_rss_LDR_ME.R
Modified: code/run_WhiteBlood_analysis_E_S_M.sh
Modified: code/run_WhiteBlood_ctwas_rss_LDR_E_S_M.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
analysis_id <- params$analysis_id
trait_id <- params$trait_id
weight <- params$weight
results_dir <- paste0("/project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/", trait_id, "/", weight)
source("/project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config_b38.R")
options(digits = 4)Load ctwas results
Check convergence of parameters
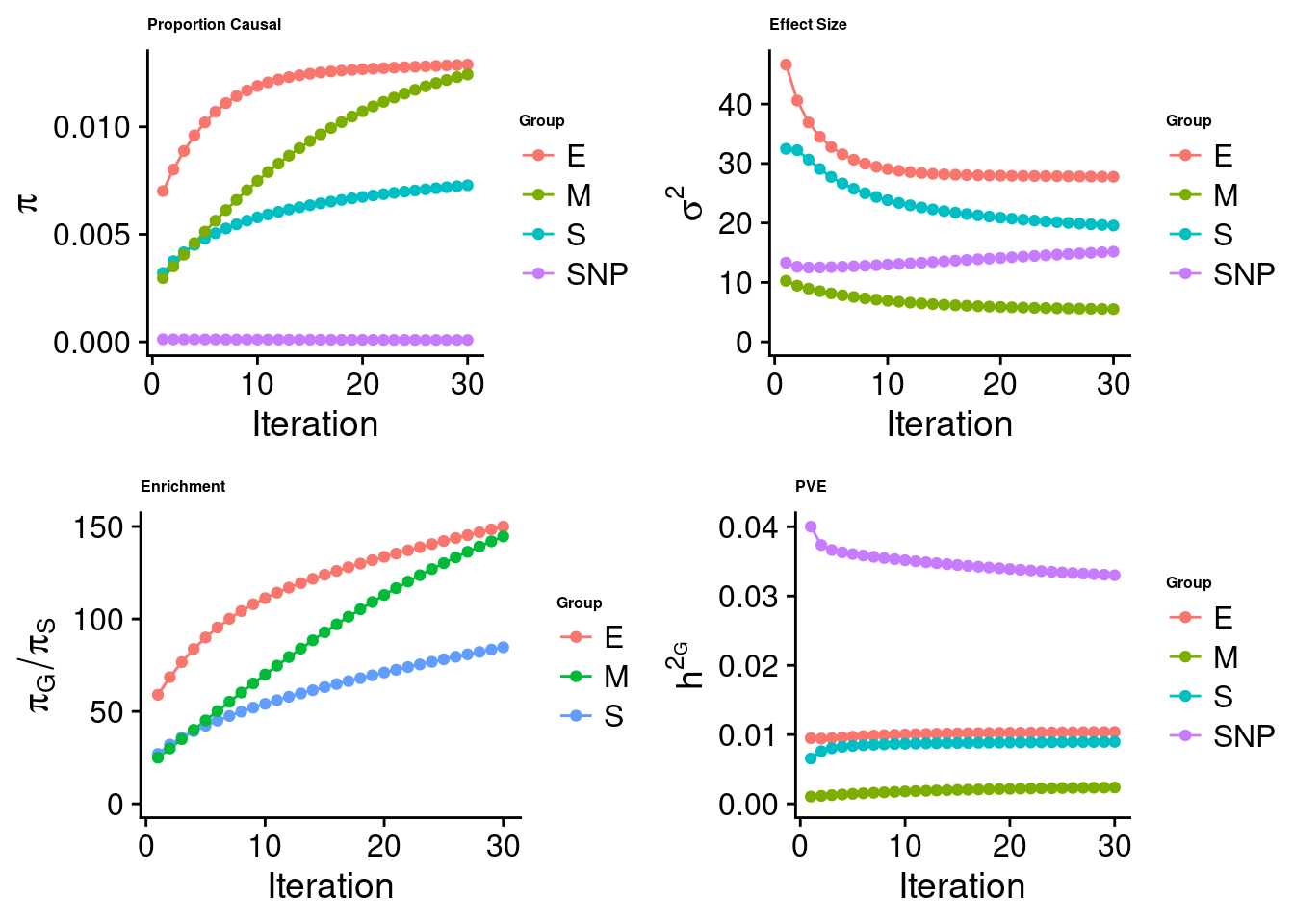
#estimated group prior
estimated_group_prior <- estimated_group_prior_all[,ncol(group_prior_rec)]
print(estimated_group_prior) SNP E S M
8.598e-05 1.289e-02 7.282e-03 1.244e-02 #estimated group prior variance
estimated_group_prior_var <- estimated_group_prior_var_all[,ncol(group_prior_var_rec)]
print(estimated_group_prior_var) SNP E S M
15.165 27.768 19.568 5.503 #estimated enrichment
estimated_enrichment <- estimated_enrichment_all[ncol(group_prior_var_rec)]
print(estimated_enrichment)[1] 69.97#report sample size
print(sample_size)[1] 343621#report group size
print(group_size) SNP E S M
8696600 9957 21596 11858 #estimated group PVE
estimated_group_pve <- estimated_group_pve_all[,ncol(group_prior_rec)]
print(estimated_group_pve) SNP E S M
0.033001 0.010375 0.008955 0.002362 #total PVE
sum(estimated_group_pve)[1] 0.05469#attributable PVE
estimated_group_pve/sum(estimated_group_pve) SNP E S M
0.60339 0.18970 0.16373 0.04318 Top expression/intron/CpG units
genename gene_id susie_pip group region_tag
42778 LDLR intron_19_11120522_11123174 1.0000 S 19_9
38222 PSRC1 ENSG00000134222.16 1.0000 E 1_67
39275 HLA-E intron_6_30490515_30491137 1.0000 S 6_24
42092 HPR ENSG00000261701.6 1.0000 E 16_38
41031 ST3GAL4 ENSG00000110080.18 1.0000 E 11_77
38537 INSIG2 ENSG00000125629.14 1.0000 E 2_69
40346 ABCA1 ENSG00000165029.15 0.9995 E 9_53
39880 NPC1L1 ENSG00000015520.14 0.9994 E 7_32
41326 GAS6 ENSG00000183087.14 0.9991 E 13_62
38489 ABCG8 ENSG00000143921.6 0.9990 E 2_27
40246 TNKS ENSG00000173273.15 0.9989 E 8_12
43341 PLTP ENSG00000100979.14 0.9989 E 20_28
39246 TRIM39 ENSG00000204599.14 0.9989 E 6_24
38587 INHBB ENSG00000163083.5 0.9977 E 2_70
40861 FADS1 ENSG00000149485.18 0.9977 E 11_34
39071 PELO ENSG00000152684.10 0.9971 E 5_30
40082 LRCH4 intron_7_100575304_100575705 0.9968 S 7_61
39103 CSNK1G3 ENSG00000151292.17 0.9963 E 5_75
42316 STAT5B ENSG00000173757.9 0.9958 E 17_25
43014 CYP2A6 ENSG00000255974.7 0.9952 E 19_28
39841 SP4 ENSG00000105866.13 0.9947 E 7_19
42113 HP intron_16_72057466_72058254 0.9940 S 16_38
43138 PRKD2 ENSG00000105287.12 0.9933 E 19_33
40798 SPTY2D1 ENSG00000179119.14 0.9888 E 11_13
38336 ALLC ENSG00000151360.9 0.9872 E 2_2
40152 POP7 ENSG00000172336.4 0.9872 E 7_62
43169 FUT2 ENSG00000176920.11 0.9860 E 19_33
38599 ACVR1C ENSG00000123612.15 0.9858 E 2_94
40593 CWF19L1 ENSG00000095485.16 0.9850 E 10_64
40754 C10orf88 ENSG00000119965.12 0.9846 E 10_77
41114 PHC1 intron_12_8921750_8922633 0.9836 S 12_9
38855 GSK3B ENSG00000082701.14 0.9831 E 3_74
42572 KDSR ENSG00000119537.15 0.9799 E 18_35
43251 RRBP1 ENSG00000125844.15 0.9773 E 20_13
38212 PRMT6 ENSG00000198890.7 0.9763 E 1_66
41831 LIPC intron_15_58431776_58431993 0.9762 S 15_26
38977 PCYT1A cg18547419 0.9722 M 3_120
40258 KIF13B intron_8_29092878_29099133 0.9671 S 8_28
42229 ATP1B2 ENSG00000129244.8 0.9656 E 17_7
39881 DDX56 ENSG00000136271.10 0.9643 E 7_32
41174 R3HDM2 intron_12_57395818_57430720 0.9616 S 12_36
42523 PGS1 intron_17_78378808_78392476 0.9581 S 17_44
40266 TTC39B ENSG00000155158.20 0.9577 E 9_13
38749 UGT1A6 intron_2_233761151_233767034 0.9574 S 2_137
41152 LRRK2 ENSG00000188906.14 0.9538 E 12_25
41476 NYNRIN ENSG00000205978.5 0.9534 E 14_3
41951 CSK cg14664628 0.9527 M 15_35
42987 CYP4F12 intron_19_15685197_15696430 0.9525 S 19_13
38009 KLHDC7A ENSG00000179023.8 0.9508 E 1_13
40386 PKN3 ENSG00000160447.6 0.9492 E 9_66
43225 SPHK2 intron_19_48624430_48625891 0.9438 S 19_33
38292 CNIH4 ENSG00000143771.11 0.9408 E 1_114
40549 SGMS1 ENSG00000198964.13 0.9383 E 10_33
43299 ZHX3 intron_20_41201384_41202057 0.9381 S 20_25
42129 OSGIN1 ENSG00000140961.12 0.9333 E 16_48
39840 DAGLB cg22939593 0.9284 M 7_8
41315 COL4A2 cg05272587 0.9092 M 13_59
40673 WBP1L ENSG00000166272.16 0.9057 E 10_66
39785 HBS1L ENSG00000112339.14 0.8972 E 6_89
39061 C4orf36 cg10685359 0.8972 M 4_59
40157 SRRT ENSG00000087087.18 0.8930 E 7_62
40216 CLDN23 ENSG00000253958.1 0.8920 E 8_11
38120 SYTL1 ENSG00000142765.17 0.8903 E 1_19
42196 ZDHHC7 cg01715842 0.8882 M 16_49
38089 ASAP3 ENSG00000088280.18 0.8851 E 1_16
39965 BRI3 ENSG00000164713.9 0.8763 E 7_60
41084 CCND2 ENSG00000118971.7 0.8735 E 12_4
40581 REEP3 cg04658946 0.8664 M 10_42
39683 TAP2 cg08327277 0.8606 M 6_27
42042 ITGAL intron_16_30505462_30506715 0.8564 S 16_24
38528 SPRED2 intron_2_65314169_65316734 0.8563 S 2_42
41627 DPF3 ENSG00000205683.11 0.8523 E 14_34
38576 CCDC93 cg02827245 0.8489 M 2_69
41617 GEMIN2 intron_14_39132068_39136440 0.8446 S 14_11
42473 ABCA8 intron_17_68883882_68884331 0.8430 S 17_39
43331 SDC4 ENSG00000124145.6 0.8365 E 20_28
40334 FOXE1 cg17702692 0.8340 M 9_49
41226 SFSWAP intron_12_131778330_131786463 0.8333 S 12_81
38007 CASZ1 cg12760995 0.8316 M 1_7
41285 TBC1D4 ENSG00000136111.12 0.8257 E 13_37
38823 FAM3D ENSG00000198643.6 0.8144 E 3_40
41812 BCL11B cg13911498 0.8116 M 14_52
38600 VIL1 ENSG00000127831.10 0.8087 E 2_129
39521 LY6G5C intron_6_31679268_31681181 0.8023 S 6_26Top genes by expression/splicing/methylation pip
genename susie_pip group
12047 HLA-E 1.7685 S
19100 PTPRN2 1.7675 M
14018 PXK 1.3158 S
14478 SEC16B 1.2167 S
11424 ERGIC3 1.1413 S
10456 CCDC57 1.1187 S
13460 PARP9 1.1092 S
10982 CYP4F12 1.0919 S
19238 RIMBP2 1.0819 M
18582 NDUFAF6 1.0768 M
14653 SLC22A18 1.0593 S
12178 IL17RC 1.0512 S
12084 HP 1.0453 S
11647 FLOT2 1.0411 S
13573 PGS1 1.0411 S
9582 ABCA8 1.0272 S
19288 RPTOR 1.0258 M
12536 LRCH4 1.0236 S
13590 PHLDB1 1.0161 S
13860 PPP6R2 1.0151 S
17100 COL4A2 1.0128 M
10037 ASGR1 1.0121 S
18823 PCYT1A 1.0103 M
12506 LIPC 1.0062 S
12462 LDLR 1.0000 S
6264 PSRC1 1.0000 E
3553 HPR 1.0000 E
7700 ST3GAL4 1.0000 E
3756 INSIG2 1.0000 E
16 ABCA1 0.9995 E
5243 NPC1L1 0.9994 E
3046 GAS6 0.9991 E
43 ABCG8 0.9990 E
8348 TNKS 0.9989 E
5928 PLTP 0.9989 E
8477 TRIM39 0.9989 E
3742 INHBB 0.9977 E
2662 FADS1 0.9977 E
5682 PELO 0.9971 E
1867 CSNK1G3 0.9963 E
7728 STAT5B 0.9958 E
1964 CYP2A6 0.9952 E
7562 SP4 0.9947 E
6162 PRKD2 0.9933 E
15728 UGT1A6 0.9921 S
7652 SPTY2D1 0.9888 E
334 ALLC 0.9872 E
6014 POP7 0.9872 E
9657 ACP6 0.9861 S
2992 FUT2 0.9860 E
137 ACVR1C 0.9858 E
1924 CWF19L1 0.9850 E
896 C10orf88 0.9846 E
13577 PHC1 0.9836 S
14918 SPRED2 0.9833 S
3322 GSK3B 0.9831 E
14040 R3HDM2 0.9820 S
3950 KDSR 0.9799 E
17745 GLIS1 0.9783 M
6839 RRBP1 0.9773 E
6169 PRMT6 0.9763 E
13063 NADK2 0.9690 S
12363 KIF13B 0.9671 S
19745 TAPBP 0.9662 M
665 ATP1B2 0.9656 E
2097 DDX56 0.9643 E
8600 TTC39B 0.9577 E
16807 CASZ1 0.9569 M
4327 LRRK2 0.9538 E
5382 NYNRIN 0.9534 E
17169 CSK 0.9527 M
4023 KLHDC7A 0.9508 E
9644 ACMSD 0.9499 S
5840 PKN3 0.9492 E
14903 SPHK2 0.9458 S
1657 CNIH4 0.9408 E
7118 SGMS1 0.9383 E
16077 ZHX3 0.9381 S
5442 OSGIN1 0.9333 E
17236 DAGLB 0.9284 M
16749 C4orf36 0.9220 M
19598 SOX30 0.9204 M
19744 TAP2 0.9160 M
8925 WBP1L 0.9057 E
3416 HBS1L 0.8972 E
7676 SRRT 0.8930 E
1579 CLDN23 0.8920 E
12272 ITGAL 0.8908 S
7871 SYTL1 0.8903 E
20147 ZDHHC7 0.8882 M
597 ASAP3 0.8851 E
15097 TACC1 0.8772 S
11642 FKRP 0.8772 S
858 BRI3 0.8763 E
11764 GEMIN2 0.8736 S
1241 CCND2 0.8735 E
13028 MYO15B 0.8722 S
14548 SFSWAP 0.8691 S
19212 REEP3 0.8664 M
2261 DPF3 0.8523 E
16856 CCDC93 0.8489 M
16655 BCL11B 0.8396 M
6990 SDC4 0.8365 E
13200 NINL 0.8364 S
17626 FOXE1 0.8340 M
7941 TBC1D4 0.8257 E
11775 GGCT 0.8202 S
2738 FAM3D 0.8144 E
8857 VIL1 0.8087 E
12590 LY6G5C 0.8023 S
13692 PLEKHA7 0.8009 STop genes by combined PIP
genename combined_pip expression_pip splicing_pip methylation_pip
4696 HLA-E 1.769 0.000 1.769 0.000
8373 PTPRN2 1.768 0.000 0.000 1.768
8400 PXK 1.675 0.337 1.316 0.023
2820 DDX56 1.565 0.964 0.601 0.000
2240 CNIH4 1.282 0.941 0.341 0.000
2714 DAGLB 1.279 0.175 0.175 0.928
9264 SEC16B 1.262 0.045 1.217 0.000
448 ALLC 1.256 0.987 0.078 0.191
6767 NDUFAF6 1.240 0.026 0.138 1.077
1633 CCDC57 1.231 0.025 1.119 0.088
7397 PARP9 1.218 0.109 1.109 0.000
2316 COL4A2 1.169 0.156 0.000 1.013
5386 KLHDC7A 1.157 0.951 0.000 0.206
3462 ERGIC3 1.141 0.000 1.141 0.000
7489 PCYT1A 1.128 0.018 0.099 1.010
9579 SLC22A18 1.127 0.000 1.059 0.067
5654 LIPC 1.113 0.107 1.006 0.000
2687 CYP4F12 1.111 0.019 1.092 0.000
8815 RIMBP2 1.102 0.020 0.000 1.082
7636 PGS1 1.100 0.027 1.041 0.032
4954 IL17RC 1.095 0.044 1.051 0.000
7574 PELO 1.093 0.997 0.096 0.000
6981 NPC1L1 1.091 0.999 0.036 0.056
9998 SP4 1.091 0.995 0.000 0.096
145 ACP6 1.088 0.102 0.986 0.000
2991 DNAJC13 1.087 0.390 0.697 0.000
9023 RPTOR 1.087 0.048 0.013 1.026
4918 IGF2R 1.076 0.282 0.794 0.000
6878 NINL 1.076 0.014 0.836 0.225
5113 ITGAL 1.074 0.000 0.891 0.183
10178 ST3GAL4 1.074 1.000 0.074 0.000
2611 CWF19L1 1.069 0.985 0.084 0.000
9031 RRBP1 1.069 0.977 0.032 0.060
3592 FADS1 1.067 0.998 0.000 0.069
9397 SGMS1 1.059 0.938 0.006 0.114
4762 HP 1.045 0.000 1.045 0.000
8110 PPP6R2 1.043 0.028 1.015 0.000
23 ABCA1 1.041 1.000 0.030 0.011
3907 FLOT2 1.041 0.000 1.041 0.000
7668 PHLDB1 1.039 0.023 1.016 0.000
30 ABCA8 1.038 0.000 1.027 0.011
807 ASAP3 1.037 0.885 0.152 0.000
11333 TTC39B 1.030 0.958 0.051 0.021
5716 LRCH4 1.029 0.005 1.024 0.000
2663 CYP2A6 1.026 0.995 0.031 0.000
9156 SAT2 1.025 0.237 0.788 0.000
11628 USP53 1.024 0.243 0.781 0.000
5295 KDSR 1.022 0.980 0.008 0.035
7643 PHC1 1.022 0.038 0.984 0.000
8434 R3HDM2 1.018 0.000 0.982 0.036
4102 GAS6 1.013 0.999 0.014 0.000
822 ASGR1 1.012 0.000 1.012 0.000
11182 TRIM39 1.010 0.999 0.011 0.000
4302 GNMT 1.008 0.052 0.210 0.746
8177 PRKD2 1.007 0.993 0.014 0.000
7883 PLTP 1.006 0.999 0.007 0.000
11520 UGT1A6 1.006 0.014 0.992 0.000
2529 CSNK1G3 1.004 0.996 0.008 0.000
1050 BCAT2 1.003 0.211 0.792 0.000
10857 TMEM199 1.001 0.651 0.350 0.000
4770 HPR 1.000 1.000 0.000 0.000
5032 INSIG2 1.000 1.000 0.000 0.000
5574 LDLR 1.000 0.000 1.000 0.000
8307 PSRC1 1.000 1.000 0.000 0.000
60 ABCG8 0.999 0.999 0.000 0.000
7468 PCMTD2 0.999 0.515 0.484 0.000
10447 TAP2 0.999 0.011 0.072 0.916
11014 TNKS 0.999 0.999 0.000 0.000
5013 INHBB 0.998 0.998 0.000 0.000
10216 STAT5B 0.996 0.996 0.000 0.000
1176 BRI3 0.993 0.876 0.117 0.000
10087 SPRED2 0.993 0.009 0.983 0.000
10448 TAPBP 0.993 0.012 0.016 0.966
10110 SPTY2D1 0.989 0.989 0.000 0.000
12066 ZHX3 0.988 0.012 0.938 0.038
7995 POP7 0.987 0.987 0.000 0.000
186 ACVR1C 0.986 0.986 0.000 0.000
4016 FUT2 0.986 0.986 0.000 0.000
1230 C10orf88 0.985 0.985 0.000 0.000
6658 NADK2 0.984 0.015 0.969 0.000
4471 GSK3B 0.983 0.983 0.000 0.000
10145 SRRT 0.981 0.893 0.088 0.000
7143 NUP88 0.980 0.046 0.528 0.405
4236 GLIS1 0.978 0.000 0.000 0.978
10054 SPHK2 0.978 0.032 0.946 0.000
8186 PRMT6 0.976 0.976 0.000 0.000
5789 LRRK2 0.974 0.954 0.020 0.000
7266 OSGIN1 0.972 0.933 0.000 0.039
11763 WBP1L 0.969 0.906 0.017 0.046
5329 KIF13B 0.967 0.000 0.967 0.000
904 ATP1B2 0.966 0.966 0.000 0.000
10665 THOP1 0.958 0.188 0.724 0.047
1529 CASZ1 0.957 0.000 0.000 0.957
2520 CSK 0.953 0.000 0.000 0.953
7162 NYNRIN 0.953 0.953 0.000 0.000
126 ACMSD 0.950 0.000 0.950 0.000
7777 PKN3 0.949 0.949 0.000 0.000
6637 N4BP2L2 0.936 0.543 0.393 0.000
1363 C4orf36 0.935 0.013 0.000 0.922
2581 CTSH 0.935 0.745 0.162 0.029
8538 RALGDS 0.932 0.000 0.605 0.327
12019 ZDHHC7 0.930 0.012 0.030 0.888
6594 MYO15B 0.925 0.015 0.872 0.037
9989 SOX30 0.920 0.000 0.000 0.920
8356 PTPN2 0.919 0.063 0.372 0.484
10398 SYTL1 0.912 0.890 0.021 0.000
4578 HBS1L 0.910 0.897 0.013 0.000
8202 PROZ 0.909 0.015 0.207 0.688
8714 REPS1 0.906 0.279 0.557 0.069
10488 TBC1D4 0.904 0.826 0.015 0.063
1936 CENPW 0.903 0.000 0.277 0.625
9378 SFSWAP 0.902 0.000 0.869 0.033
3058 DPF3 0.900 0.852 0.000 0.048
4158 GEMIN2 0.900 0.026 0.874 0.000
2147 CLDN23 0.892 0.892 0.000 0.000
10408 TACC1 0.890 0.012 0.877 0.000
1661 CCDC93 0.884 0.009 0.026 0.849
8461 RAB2A 0.883 0.125 0.038 0.720
3897 FKRP 0.882 0.005 0.877 0.000
8701 REEP3 0.882 0.009 0.007 0.866
1692 CCND2 0.873 0.873 0.000 0.000
7697 PIEZO1 0.869 0.060 0.125 0.683
3859 FGFR2 0.867 0.030 0.052 0.785
8094 PPP2R5C 0.866 0.067 0.799 0.000
9980 SORCS2 0.859 0.753 0.013 0.093
5839 LY6G5C 0.844 0.008 0.802 0.034
9232 SDC4 0.843 0.837 0.006 0.000
4178 GGCT 0.841 0.021 0.820 0.000
6877 NINJ2 0.841 0.022 0.311 0.508
1058 BCL11B 0.840 0.000 0.000 0.840
2422 CPNE3 0.840 0.141 0.000 0.699
3955 FOXE1 0.834 0.000 0.000 0.834
8018 PPCDC 0.824 0.460 0.323 0.040
9536 SLC12A7 0.823 0.059 0.336 0.429
11681 VIL1 0.823 0.809 0.014 0.000
7824 PLEC 0.822 0.027 0.025 0.769
7833 PLEKHA7 0.821 0.020 0.801 0.000
6540 MVB12A 0.818 0.718 0.101 0.000
3689 FAM3D 0.814 0.814 0.000 0.000
11758 WASHC3 0.813 0.027 0.786 0.000
232 ADARB2 0.811 0.013 0.000 0.798
10498 TBCD 0.802 0.022 0.138 0.641
region_tag
4696 6_24
8373 7_98
8400 3_40
2820 7_32
2240 1_114
2714 7_8
9264 1_87
448 2_2
6767 8_66
1633 17_47
7397 3_76
2316 13_59
5386 1_13
3462 20_21
7489 3_121
9579 11_2
5654 15_26
2687 19_13
8815 12_80
7636 17_44
4954 3_8
7574 5_30
6981 7_32
9998 7_19
145 1_73
2991 3_82
9023 17_45
4918 6_103
6878 20_19
5113 16_24
10178 11_77
2611 10_64
9031 20_13
3592 11_34
9397 10_33
4762 16_38
8110 22_24
23 9_53
3907 17_17
7668 11_71
30 17_39
807 1_16
11333 9_13
5716 7_61
2663 19_28
9156 17_7
11628 4_77
5295 18_35
7643 12_9
8434 12_36
4102 13_62
822 17_6
11182 6_24
4302 6_33
8177 19_33
7883 20_28
11520 2_137
2529 5_75
1050 19_34
10857 17_17
4770 16_38
5032 2_69
5574 19_9
8307 1_67
60 2_27
7468 20_38
10447 6_27
11014 8_12
5013 2_70
10216 17_25
1176 7_60
10087 2_42
10448 6_28
10110 11_13
12066 20_25
7995 7_62
186 2_94
4016 19_33
1230 10_77
6658 5_24
4471 3_74
10145 7_62
7143 17_5
4236 1_33
10054 19_33
8186 1_66
5789 12_25
7266 16_48
11763 10_66
5329 8_28
904 17_7
10665 19_3
1529 1_7
2520 15_35
7162 14_3
126 2_79
7777 9_66
6637 13_10
1363 4_59
2581 15_37
8538 9_70
12019 16_49
6594 17_42
9989 5_93
8356 18_9
10398 1_19
4578 6_89
8202 13_62
8714 6_92
10488 13_37
1936 6_84
9378 12_81
3058 14_34
4158 14_11
2147 8_11
10408 8_34
1661 2_69
8461 8_46
3897 19_33
8701 10_42
1692 12_4
7697 16_53
3859 10_76
8094 14_53
9980 4_8
5839 6_26
9232 20_28
4178 7_24
6877 12_1
1058 14_52
2422 8_62
3955 9_49
8018 15_35
9536 5_2
11681 2_129
7824 8_94
7833 11_12
6540 19_14
3689 3_40
11758 12_61
232 10_2
10498 17_47#pdf(file = "./LDL_TEME199_genetrack.pdf", width = 3.86, height = 0.4)
#locus_plot_gene_track_pub(a, label_pos="above")
#dev.off()
sessionInfo()R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] cowplot_1.1.1 ggplot2_3.4.0 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] tidyselect_1.2.0 xfun_0.35 bslib_0.4.1 colorspace_2.0-3
[5] vctrs_0.5.1 generics_0.1.3 htmltools_0.5.4 yaml_2.3.6
[9] blob_1.2.3 utf8_1.2.2 rlang_1.0.6 jquerylib_0.1.4
[13] later_1.3.0 pillar_1.8.1 glue_1.6.2 withr_2.5.0
[17] DBI_1.1.3 bit64_4.0.5 lifecycle_1.0.3 stringr_1.5.0
[21] munsell_0.5.0 gtable_0.3.1 memoise_2.0.1 evaluate_0.19
[25] labeling_0.4.2 knitr_1.41 callr_3.7.3 fastmap_1.1.0
[29] httpuv_1.6.7 ps_1.7.2 fansi_1.0.3 highr_0.9
[33] Rcpp_1.0.9 promises_1.2.0.1 scales_1.2.1 cachem_1.0.6
[37] jsonlite_1.8.4 bit_4.0.5 farver_2.1.0 fs_1.5.2
[41] digest_0.6.31 stringi_1.7.8 processx_3.8.0 dplyr_1.0.10
[45] getPass_0.2-2 rprojroot_2.0.3 grid_4.1.0 cli_3.4.1
[49] tools_4.1.0 magrittr_2.0.3 sass_0.4.4 RSQLite_2.2.19
[53] tibble_3.1.8 whisker_0.4.1 pkgconfig_2.0.3 data.table_1.14.6
[57] assertthat_0.2.1 rmarkdown_2.19 httr_1.4.4 rstudioapi_0.14
[61] R6_2.5.1 git2r_0.30.1 compiler_4.1.0