LDL - Liver
sheng Qian
2022-10-18
Last updated: 2023-03-30
Checks: 5 2
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config_b38.R | code/ctwas_config_b38.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 4c76b6a. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .ipynb_checkpoints/
Untracked files:
Untracked: LDL_LDLR_genetrack.pdf
Untracked: LDL_LDLR_locus.pdf
Untracked: LDL_TEME199_genetrack.pdf
Untracked: LDL_TEME199_locus.pdf
Untracked: Proposal plots.R
Untracked: RGS14.pdf
Untracked: RNF186.pdf
Untracked: Rplots.pdf
Untracked: SCZ_annotation.xlsx
Untracked: SLC8B1.pdf
Untracked: analysis/.ipynb_checkpoints/
Untracked: cache/
Untracked: code/.ipynb_checkpoints/
Untracked: data/.ipynb_checkpoints/
Untracked: data/FUMA_output/
Untracked: data/GO_Terms/
Untracked: data/GTEx_Analysis_v8_eQTL.tar
Untracked: data/G_list.RData
Untracked: data/IBD_ME/
Untracked: data/LDL/
Untracked: data/LDL_03/
Untracked: data/LDL_05/
Untracked: data/LDL_E_S/
Untracked: data/LDL_M/
Untracked: data/LDL_S/
Untracked: data/PGC3_SCZ_wave3_public.v2.tsv
Untracked: data/SCZ/
Untracked: data/SCZ_2018/
Untracked: data/SCZ_2018_S/
Untracked: data/SCZ_2020/
Untracked: data/SCZ_S/
Untracked: data/Supplementary Table 15 - MAGMA.xlsx
Untracked: data/Supplementary Table 20 - Prioritised Genes.xlsx
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/WhiteBlood_E/
Untracked: data/WhiteBlood_E_M/
Untracked: data/WhiteBlood_E_S_M/
Untracked: data/WhiteBlood_M/
Untracked: data/cpg_annot.RData
Untracked: data/eqtl/
Untracked: data/gencode.v26.GRCh38.genes.gtf
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/gwas_sumstats/
Untracked: data/magma.genes.out
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/mqtl/
Untracked: data/notes.txt
Untracked: data/scz_2018.RDS
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: hist.pdf
Untracked: submit.sh
Untracked: temp.regionlist.RDS
Untracked: temp.regions.txt
Untracked: temp.susieIrss.txt
Untracked: temp.temp.susieIrssres.Rd
Untracked: temp_LDR/
Untracked: temp_ld_R_chr1.txt
Untracked: temp_ld_R_chr10.txt
Untracked: temp_ld_R_chr11.txt
Untracked: temp_ld_R_chr12.txt
Untracked: temp_ld_R_chr13.txt
Untracked: temp_ld_R_chr14.txt
Untracked: temp_ld_R_chr15.txt
Untracked: temp_ld_R_chr16.txt
Untracked: temp_ld_R_chr17.txt
Untracked: temp_ld_R_chr18.txt
Untracked: temp_ld_R_chr19.txt
Untracked: temp_ld_R_chr2.txt
Untracked: temp_ld_R_chr20.txt
Untracked: temp_ld_R_chr21.txt
Untracked: temp_ld_R_chr22.txt
Untracked: temp_ld_R_chr3.txt
Untracked: temp_ld_R_chr4.txt
Untracked: temp_ld_R_chr5.txt
Untracked: temp_ld_R_chr6.txt
Untracked: temp_ld_R_chr7.txt
Untracked: temp_ld_R_chr8.txt
Untracked: temp_ld_R_chr9.txt
Untracked: temp_reg.txt
Unstaged changes:
Deleted: analysis/Atrial_Fibrillation_Heart_Atrial_Appendage.Rmd
Deleted: analysis/Atrial_Fibrillation_Heart_Left_Ventricle.Rmd
Deleted: analysis/Autism_Brain_Amygdala.Rmd
Deleted: analysis/Autism_Brain_Anterior_cingulate_cortex_BA24.Rmd
Deleted: analysis/Autism_Brain_Caudate_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Cerebellar_Hemisphere.Rmd
Deleted: analysis/Autism_Brain_Cerebellum.Rmd
Deleted: analysis/Autism_Brain_Cortex.Rmd
Deleted: analysis/Autism_Brain_Frontal_Cortex_BA9.Rmd
Deleted: analysis/Autism_Brain_Hippocampus.Rmd
Deleted: analysis/Autism_Brain_Hypothalamus.Rmd
Deleted: analysis/Autism_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Putamen_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Spinal_cord_cervical_c-1.Rmd
Deleted: analysis/Autism_Brain_Substantia_nigra.Rmd
Deleted: analysis/BMI_Brain_Amygdala.Rmd
Deleted: analysis/BMI_Brain_Amygdala_S.Rmd
Deleted: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24.Rmd
Deleted: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24_S.Rmd
Deleted: analysis/BMI_Brain_Caudate_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Caudate_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Cerebellar_Hemisphere.Rmd
Deleted: analysis/BMI_Brain_Cerebellar_Hemisphere_S.Rmd
Deleted: analysis/BMI_Brain_Cerebellum.Rmd
Deleted: analysis/BMI_Brain_Cerebellum_S.Rmd
Deleted: analysis/BMI_Brain_Cortex.Rmd
Deleted: analysis/BMI_Brain_Cortex_S.Rmd
Deleted: analysis/BMI_Brain_Frontal_Cortex_BA9.Rmd
Deleted: analysis/BMI_Brain_Frontal_Cortex_BA9_S.Rmd
Deleted: analysis/BMI_Brain_Hippocampus.Rmd
Deleted: analysis/BMI_Brain_Hippocampus_S.Rmd
Deleted: analysis/BMI_Brain_Hypothalamus.Rmd
Deleted: analysis/BMI_Brain_Hypothalamus_S.Rmd
Deleted: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Putamen_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Putamen_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Spinal_cord_cervical_c-1.Rmd
Deleted: analysis/BMI_Brain_Spinal_cord_cervical_c-1_S.Rmd
Deleted: analysis/BMI_Brain_Substantia_nigra.Rmd
Deleted: analysis/BMI_Brain_Substantia_nigra_S.Rmd
Deleted: analysis/BMI_S_results.Rmd
Deleted: analysis/Glucose_Adipose_Subcutaneous.Rmd
Deleted: analysis/Glucose_Adipose_Visceral_Omentum.Rmd
Modified: analysis/LDL_Liver_03.Rmd
Modified: analysis/LDL_Liver_05.Rmd
Deleted: code/White_Blood_M_out/White_Blood_BreastMammary.err
Deleted: code/White_Blood_M_out/White_Blood_BreastMammary.out
Deleted: code/White_Blood_M_out/White_Blood_ColonTransverse.err
Deleted: code/White_Blood_M_out/White_Blood_ColonTransverse.out
Deleted: code/White_Blood_M_out/White_Blood_KidneyCortex.err
Deleted: code/White_Blood_M_out/White_Blood_KidneyCortex.out
Deleted: code/White_Blood_M_out/White_Blood_Lung.err
Deleted: code/White_Blood_M_out/White_Blood_Lung.out
Deleted: code/White_Blood_M_out/White_Blood_MuscleSkeletal.err
Deleted: code/White_Blood_M_out/White_Blood_MuscleSkeletal.out
Deleted: code/White_Blood_M_out/White_Blood_Ovary.err
Deleted: code/White_Blood_M_out/White_Blood_Ovary.out
Deleted: code/White_Blood_M_out/White_Blood_Prostate.err
Deleted: code/White_Blood_M_out/White_Blood_Prostate.out
Deleted: code/White_Blood_M_out/White_Blood_Testis.err
Deleted: code/White_Blood_M_out/White_Blood_Testis.out
Deleted: code/White_Blood_M_out/White_Blood_WholeBlood.err
Deleted: code/White_Blood_M_out/White_Blood_WholeBlood.out
Deleted: code/run_IBD_ctwas_rss_LDR_ME.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/LDL_Liver_03.Rmd) and HTML (docs/LDL_Liver_03.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 4c76b6a | sq-96 | 2023-03-30 | update |
| html | 4c76b6a | sq-96 | 2023-03-30 | update |
Weight QC
[1] 12714[1] 10901
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
1070 768 652 417 494 611 548 408 405 434 634 629 195 365 354 526
17 18 19 20 21 22
663 160 859 306 114 289 [1] 0.8365Load ctwas results
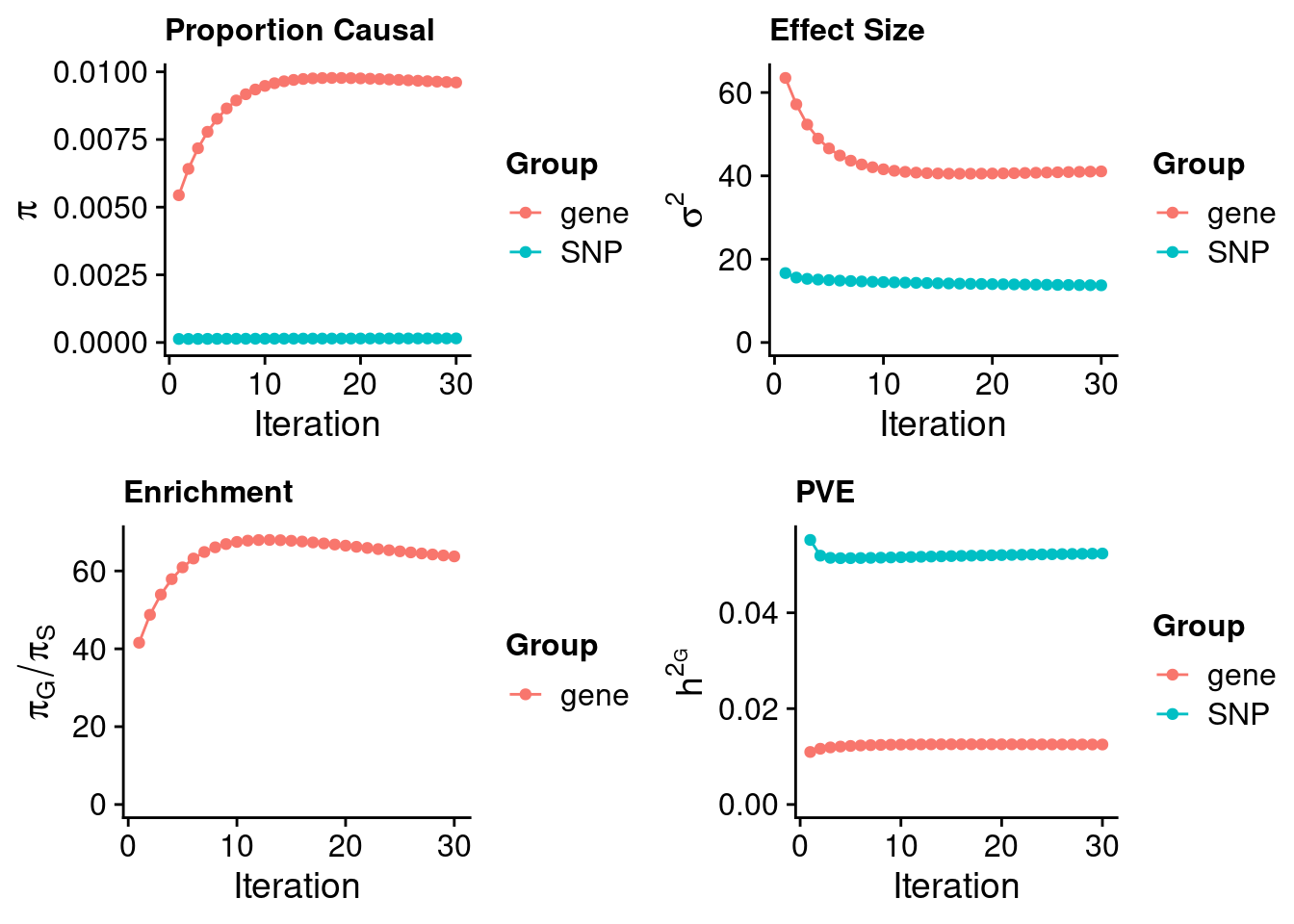
Check convergence of parameters
| Version | Author | Date |
|---|---|---|
| 4c76b6a | sq-96 | 2023-03-30 |
#estimated group prior
estimated_group_prior <- estimated_group_prior_all[,ncol(group_prior_rec)]
print(estimated_group_prior) SNP gene
0.0001507 0.0096062 #estimated group prior variance
estimated_group_prior_var <- estimated_group_prior_var_all[,ncol(group_prior_var_rec)]
print(estimated_group_prior_var) SNP gene
13.73 41.07 #estimated enrichment
estimated_enrichment <- estimated_enrichment_all[ncol(group_prior_var_rec)]
print(estimated_enrichment)[1] 63.74#report sample size
print(sample_size)[1] 343621#report group size
print(group_size) SNP gene
8696847 10901 #estimated group PVE
estimated_group_pve <- estimated_group_pve_all[,ncol(group_prior_rec)]
print(estimated_group_pve) SNP gene
0.05237 0.01252 #total PVE
sum(estimated_group_pve)[1] 0.06488#attributable PVE
estimated_group_pve/sum(estimated_group_pve) SNP gene
0.8071 0.1929 Genes with highest PIPs
#distribution of PIPs
hist(ctwas_gene_res$susie_pip, xlim=c(0,1), main="Distribution of Gene PIPs")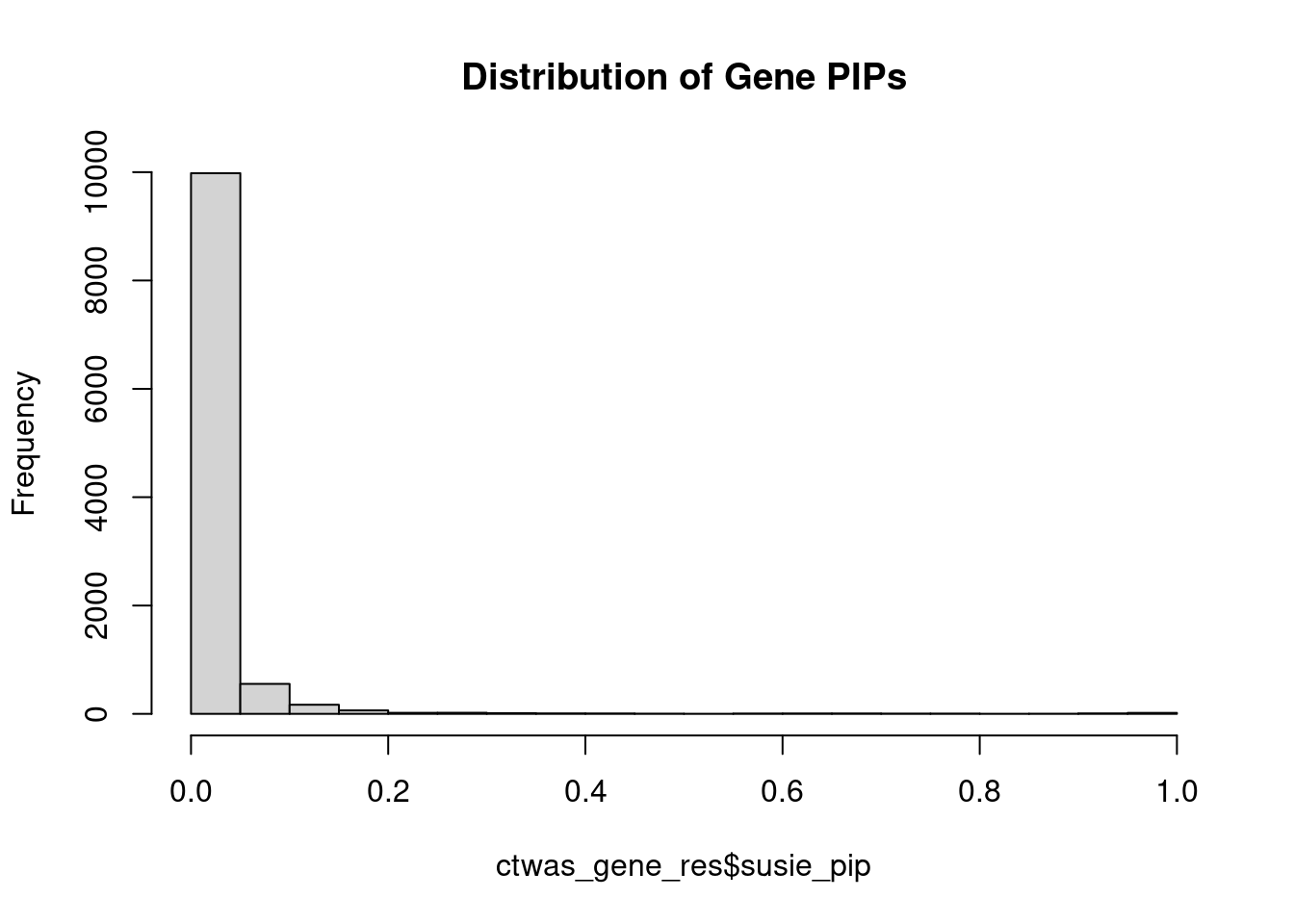
| Version | Author | Date |
|---|---|---|
| 4c76b6a | sq-96 | 2023-03-30 |
#genes with PIP>0.8 or 20 highest PIPs
head(ctwas_gene_res[order(-ctwas_gene_res$susie_pip),report_cols], max(sum(ctwas_gene_res$susie_pip>0.8), 20)) genename region_tag susie_pip mu2 PVE z num_eqtl
4435 PSRC1 1_67 1.0000 1668.11 4.854e-03 -41.687 1
12008 HPR 16_38 1.0000 158.39 4.609e-04 -17.963 2
3721 INSIG2 2_69 1.0000 68.27 1.987e-04 -8.983 3
5991 FADS1 11_34 0.9995 163.37 4.752e-04 12.926 2
7997 TMEM150A 2_54 0.9994 849.53 2.471e-03 4.079 1
5563 ABCG8 2_27 0.9980 310.09 9.006e-04 -20.294 1
10657 TRIM39 6_24 0.9975 71.45 2.074e-04 8.840 3
12687 RP4-781K5.7 1_121 0.9967 202.33 5.869e-04 -15.108 1
7410 ABCA1 9_53 0.9920 70.21 2.027e-04 7.982 1
1999 PRKD2 19_33 0.9871 29.98 8.613e-05 5.072 2
1597 PLTP 20_28 0.9827 62.58 1.790e-04 -5.732 1
8531 TNKS 8_12 0.9820 74.84 2.139e-04 11.039 2
9390 GAS6 13_62 0.9779 71.01 2.021e-04 -8.924 1
5544 CNIH4 1_114 0.9761 40.67 1.155e-04 6.146 2
7040 INHBB 2_70 0.9651 73.84 2.074e-04 -8.519 1
11790 CYP2A6 19_28 0.9590 31.93 8.910e-05 5.407 1
3247 KDSR 18_35 0.9570 24.60 6.851e-05 -4.526 1
6391 TTC39B 9_13 0.9516 23.08 6.391e-05 -4.334 3
6093 CSNK1G3 5_75 0.9469 83.85 2.311e-04 9.116 1
1114 SRRT 7_62 0.9386 32.69 8.929e-05 5.425 2
2092 SP4 7_19 0.9349 101.60 2.764e-04 10.693 1
8579 STAT5B 17_25 0.9298 30.76 8.324e-05 5.426 2
6778 PKN3 9_66 0.9277 47.46 1.281e-04 -6.621 1
3300 C10orf88 10_77 0.9269 36.76 9.916e-05 -6.788 2
3562 ACVR1C 2_94 0.9266 25.82 6.963e-05 -4.687 2
4704 DDX56 7_32 0.9161 58.85 1.569e-04 9.642 2
8865 FUT2 19_33 0.9022 104.08 2.733e-04 -11.927 1
6957 USP1 1_39 0.8889 253.52 6.558e-04 16.258 1
6220 PELO 5_31 0.8758 70.31 1.792e-04 8.288 2
9062 KLHDC7A 1_13 0.8541 21.86 5.434e-05 4.124 1
5415 SYTL1 1_19 0.8315 21.89 5.296e-05 -3.963 1
8931 CRACR2B 11_1 0.8189 21.62 5.151e-05 -3.990 1GO enrichment analysis for genes with PIP>0.8
#number of genes for gene set enrichment
length(genes)[1] 32DisGeNET enrichment analysis for genes with PIP>0.5
Description FDR Ratio BgRatio
5 Blood Platelet Disorders 0.01102 2/16 16/9703
23 Hypercholesterolemia, Familial 0.01102 2/16 18/9703
30 Leukemia, T-Cell, Chronic 0.01102 1/16 1/9703
39 Opisthorchiasis 0.01102 1/16 1/9703
47 Tangier Disease 0.01102 1/16 1/9703
63 Caliciviridae Infections 0.01102 1/16 1/9703
69 Infections, Calicivirus 0.01102 1/16 1/9703
83 Opisthorchis felineus Infection 0.01102 1/16 1/9703
84 Opisthorchis viverrini Infection 0.01102 1/16 1/9703
97 Enteropathy-Associated T-Cell Lymphoma 0.01102 1/16 1/9703WebGestalt enrichment analysis for genes with PIP>0.5
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...
description size overlap FDR database
1 Dyslipidaemia 84 7 2.712e-05 disease_GLAD4U
2 Coronary Artery Disease 153 8 4.279e-05 disease_GLAD4U
3 Coronary Disease 171 8 6.796e-05 disease_GLAD4U
4 Arteriosclerosis 173 7 9.859e-04 disease_GLAD4U
5 Myocardial Ischemia 181 7 1.068e-03 disease_GLAD4U
6 Arterial Occlusive Diseases 174 6 9.988e-03 disease_GLAD4U
7 Cardiovascular Diseases 282 7 1.392e-02 disease_GLAD4U
8 Hypercholesterolemia 60 4 1.499e-02 disease_GLAD4U
9 Heart Diseases 227 6 2.944e-02 disease_GLAD4U
10 Cholesterol metabolism 31 3 3.711e-02 pathway_KEGG
userId
1 PSRC1;ABCG8;INSIG2;TTC39B;ABCA1;FADS1;PLTP
2 PSRC1;ABCG8;INSIG2;TTC39B;ABCA1;FADS1;FUT2;PLTP
3 PSRC1;ABCG8;INSIG2;TTC39B;ABCA1;FADS1;FUT2;PLTP
4 PSRC1;ABCG8;TTC39B;ABCA1;FADS1;HPR;PLTP
5 PSRC1;ABCG8;INSIG2;TTC39B;ABCA1;FADS1;PLTP
6 PSRC1;ABCG8;TTC39B;ABCA1;FADS1;PLTP
7 PSRC1;ABCG8;TTC39B;ABCA1;FADS1;GAS6;PLTP
8 ABCG8;INSIG2;ABCA1;PLTP
9 PSRC1;ABCG8;TTC39B;ABCA1;FADS1;PLTP
10 ABCG8;ABCA1;PLTP
sessionInfo()R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] WebGestaltR_0.4.4 disgenet2r_0.99.2 enrichR_3.1 cowplot_1.1.1
[5] ggplot2_3.4.0 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] httr_1.4.4 sass_0.4.4 vroom_1.6.0 bit64_4.0.5
[5] jsonlite_1.8.4 foreach_1.5.2 bslib_0.4.1 assertthat_0.2.1
[9] getPass_0.2-2 highr_0.9 doRNG_1.8.2 blob_1.2.3
[13] yaml_2.3.6 pillar_1.8.1 RSQLite_2.2.19 lattice_0.20-44
[17] glue_1.6.2 digest_0.6.31 promises_1.2.0.1 colorspace_2.0-3
[21] htmltools_0.5.4 httpuv_1.6.7 Matrix_1.3-3 plyr_1.8.8
[25] pkgconfig_2.0.3 scales_1.2.1 svglite_2.1.0 processx_3.8.0
[29] whisker_0.4.1 later_1.3.0 tzdb_0.3.0 git2r_0.30.1
[33] tibble_3.1.8 generics_0.1.3 farver_2.1.0 ellipsis_0.3.2
[37] cachem_1.0.6 withr_2.5.0 cli_3.4.1 crayon_1.5.2
[41] magrittr_2.0.3 memoise_2.0.1 evaluate_0.19 ps_1.7.2
[45] apcluster_1.4.10 fs_1.5.2 fansi_1.0.3 doParallel_1.0.17
[49] tools_4.1.0 data.table_1.14.6 hms_1.1.2 lifecycle_1.0.3
[53] stringr_1.5.0 munsell_0.5.0 rngtools_1.5.2 callr_3.7.3
[57] compiler_4.1.0 jquerylib_0.1.4 systemfonts_1.0.4 rlang_1.0.6
[61] grid_4.1.0 iterators_1.0.14 rstudioapi_0.14 rjson_0.2.21
[65] igraph_1.3.5 labeling_0.4.2 rmarkdown_2.19 gtable_0.3.1
[69] codetools_0.2-18 DBI_1.1.3 curl_4.3.2 reshape2_1.4.4
[73] R6_2.5.1 knitr_1.41 dplyr_1.0.10 fastmap_1.1.0
[77] bit_4.0.5 utf8_1.2.2 rprojroot_2.0.3 readr_2.1.3
[81] stringi_1.7.8 parallel_4.1.0 Rcpp_1.0.9 vctrs_0.5.1
[85] tidyselect_1.2.0 xfun_0.35