BMI - Brain Anterior cingulate cortex BA24
sheng Qian
2021-2-6
Last updated: 2022-02-21
Checks: 6 1
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version bbf6737. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Untracked files:
Untracked: Rplot.png
Untracked: analysis/Glucose_Adipose_Subcutaneous.Rmd
Untracked: analysis/Glucose_Adipose_Visceral_Omentum.Rmd
Untracked: analysis/Splicing_Test.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/BMI_S_out/
Untracked: code/BMI_out/
Untracked: code/Glucose_out/
Untracked: code/LDL_S_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_analysis_S.sbatch
Untracked: code/run_BMI_analysis_S.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_BMI_ctwas_rss_LDR_S.R
Untracked: code/run_Glucose_analysis.sbatch
Untracked: code/run_Glucose_analysis.sh
Untracked: code/run_Glucose_ctwas_rss_LDR.R
Untracked: code/run_LDL_analysis_S.sbatch
Untracked: code/run_LDL_analysis_S.sh
Untracked: code/run_LDL_ctwas_rss_LDR_S.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: data/.ipynb_checkpoints/
Untracked: data/AF/
Untracked: data/BMI/
Untracked: data/BMI_S/
Untracked: data/Glucose/
Untracked: data/LDL_S/
Untracked: data/T2D/
Untracked: data/TEST/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/BMI_Brain_Anterior_cingulate_cortex_BA24.Rmd) and HTML (docs/BMI_Brain_Anterior_cingulate_cortex_BA24.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | bbf6737 | sq-96 | 2022-02-21 | update |
| html | 91f38fa | sq-96 | 2022-02-13 | Build site. |
| Rmd | eb13ecf | sq-96 | 2022-02-13 | update |
| html | e6bc169 | sq-96 | 2022-02-13 | Build site. |
| Rmd | 87fee8b | sq-96 | 2022-02-13 | update |
Weight QC
#number of imputed weights
nrow(qclist_all)[1] 10914#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
1077 759 640 423 520 617 523 418 401 429 656 600 225 367 374 510
17 18 19 20 21 22
639 170 840 329 119 278 #number of imputed weights without missing variants
sum(qclist_all$nmiss==0)[1] 8792#proportion of imputed weights without missing variants
mean(qclist_all$nmiss==0)[1] 0.8056Check convergence of parameters

| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
#estimated group prior
estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
names(estimated_group_prior) <- c("gene", "snp")
estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
print(estimated_group_prior) gene snp
0.0075152 0.0002941 #estimated group prior variance
estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
names(estimated_group_prior_var) <- c("gene", "snp")
print(estimated_group_prior_var) gene snp
20.14 17.22 #report sample size
print(sample_size)[1] 336107#report group size
group_size <- c(nrow(ctwas_gene_res), n_snps)
print(group_size)[1] 10914 7535010#estimated group PVE
estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size #check PVE calculation
names(estimated_group_pve) <- c("gene", "snp")
print(estimated_group_pve) gene snp
0.004914 0.113548 #compare sum(PIP*mu2/sample_size) with above PVE calculation
c(sum(ctwas_gene_res$PVE),sum(ctwas_snp_res$PVE))[1] 0.1775 15.9888Genes with highest PIPs
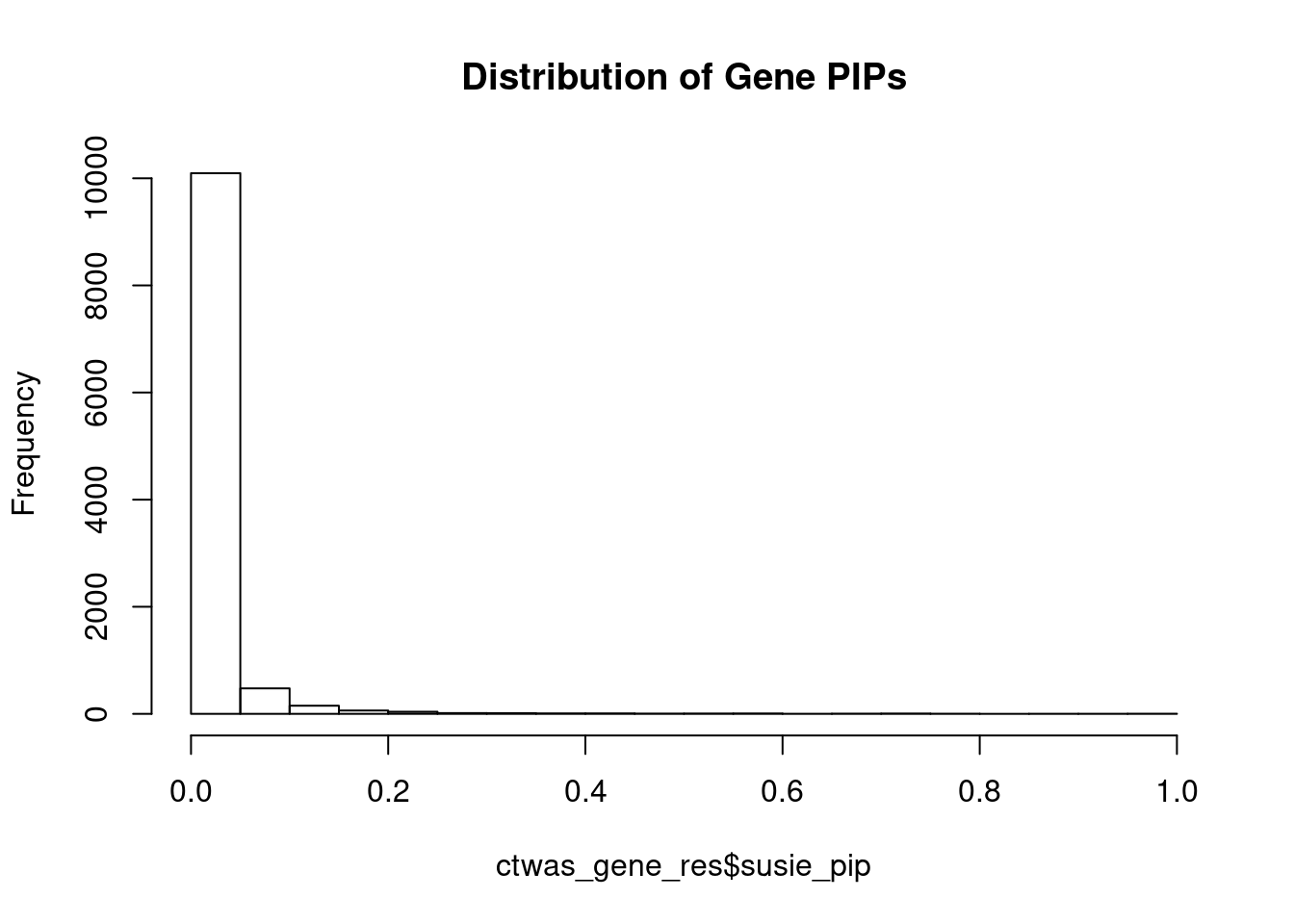
genename region_tag susie_pip mu2 PVE z num_eqtl
8261 MRPL1 4_52 1.0000 18758.93 5.581e-02 3.486 1
12013 CCDC169 13_13 1.0000 11401.83 3.392e-02 -2.791 1
2884 ARHGEF26 3_95 1.0000 20272.04 6.031e-02 -4.171 1
925 PIK3C3 18_23 0.9334 52.04 1.445e-04 6.867 2
12058 ETV5 3_114 0.8937 95.62 2.543e-04 9.862 1
9400 SHARPIN 8_94 0.8589 25.31 6.467e-05 -4.431 1
2457 STIM2 4_22 0.7911 23.30 5.483e-05 -4.484 2
4656 HTR1B 6_53 0.7783 26.47 6.130e-05 -4.637 1
3541 ZMIZ2 7_33 0.7565 66.00 1.486e-04 -8.105 1
9452 TH 11_2 0.7321 27.23 5.931e-05 4.799 1
9668 KCNB2 8_53 0.7286 65.38 1.417e-04 -8.226 1
279 CPS1 2_124 0.7268 4573.07 9.889e-03 -3.535 1
5792 ECE2 3_113 0.7217 29.17 6.263e-05 -5.315 1
9221 PNPLA2 11_1 0.7210 26.40 5.664e-05 5.118 1
318 VRK2 2_38 0.7086 24.10 5.080e-05 3.527 2
12683 LINC01977 17_45 0.6965 28.35 5.875e-05 5.230 1
7428 PPM1M 3_37 0.6866 185.89 3.797e-04 4.213 2
10559 SRGAP1 12_39 0.6839 23.33 4.747e-05 -4.065 2
7744 R3HCC1L 10_62 0.6724 39.81 7.965e-05 7.439 1
10515 ZKSCAN5 7_61 0.6438 53.56 1.026e-04 7.133 1Genes with largest effect sizes
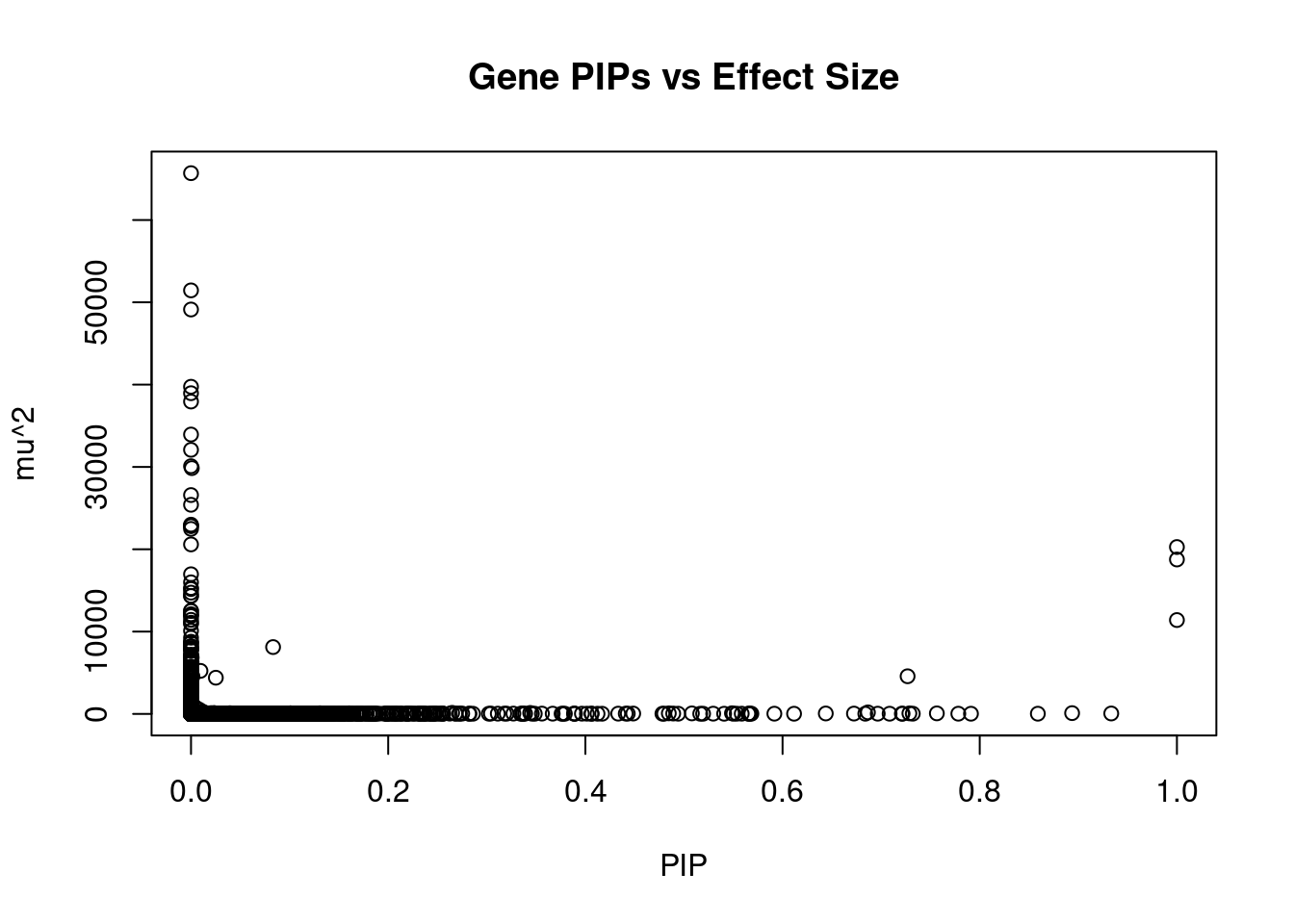
genename region_tag susie_pip mu2 PVE z num_eqtl
10292 SLC38A3 3_35 0.000e+00 65690 0.000e+00 6.726 1
7423 CAMKV 3_35 0.000e+00 51451 0.000e+00 -9.848 1
7600 CCDC171 9_13 0.000e+00 49127 0.000e+00 8.020 2
38 RBM6 3_35 0.000e+00 39743 0.000e+00 12.536 1
9 SEMA3F 3_35 0.000e+00 38958 0.000e+00 12.511 1
11183 DEFB134 8_15 0.000e+00 37947 0.000e+00 -6.778 1
7425 MST1R 3_35 0.000e+00 33940 0.000e+00 -12.621 2
4697 HEY2 6_84 0.000e+00 32081 0.000e+00 4.923 1
9326 STX19 3_59 0.000e+00 30090 0.000e+00 -5.060 1
12530 RP11-490G2.2 1_60 8.154e-04 29861 7.245e-05 5.044 1
7824 LEO1 15_21 1.925e-11 26586 1.523e-12 4.647 1
5275 LYSMD2 15_21 0.000e+00 25404 0.000e+00 -4.403 1
5271 MFAP1 15_16 0.000e+00 22989 0.000e+00 4.303 1
11993 HYPK 15_16 0.000e+00 22890 0.000e+00 4.322 1
9321 DHFR2 3_59 0.000e+00 22806 0.000e+00 3.751 2
7420 RNF123 3_35 0.000e+00 22473 0.000e+00 -10.959 1
11448 CKMT1A 15_16 0.000e+00 20590 0.000e+00 4.130 1
2884 ARHGEF26 3_95 1.000e+00 20272 6.031e-02 -4.171 1
8261 MRPL1 4_52 1.000e+00 18759 5.581e-02 3.486 1
5134 CNOT6L 4_52 0.000e+00 16966 0.000e+00 3.413 1Genes with highest PVE
genename region_tag susie_pip mu2 PVE z num_eqtl
2884 ARHGEF26 3_95 0.9999995 20272.04 6.031e-02 -4.171 1
8261 MRPL1 4_52 1.0000000 18758.93 5.581e-02 3.486 1
12013 CCDC169 13_13 1.0000000 11401.83 3.392e-02 -2.791 1
279 CPS1 2_124 0.7268233 4573.07 9.889e-03 -3.535 1
9576 C3orf58 3_88 0.0832453 8119.53 2.011e-03 2.514 1
7428 PPM1M 3_37 0.6865923 185.89 3.797e-04 4.213 2
7455 TMEM161B 5_52 0.0251788 4397.99 3.295e-04 -8.072 2
12058 ETV5 3_114 0.8936853 95.62 2.543e-04 9.862 1
12992 CTC-498M16.4 5_52 0.0095883 5230.51 1.492e-04 7.700 2
3541 ZMIZ2 7_33 0.7564988 66.00 1.486e-04 -8.105 1
925 PIK3C3 18_23 0.9333936 52.04 1.445e-04 6.867 2
9668 KCNB2 8_53 0.7286468 65.38 1.417e-04 -8.226 1
6613 GPR61 1_67 0.5486704 79.17 1.292e-04 8.755 1
5135 USO1 4_51 0.3440095 118.69 1.215e-04 -2.134 1
10515 ZKSCAN5 7_61 0.6438389 53.56 1.026e-04 7.133 1
7849 MC4R 18_33 0.2648613 128.46 1.012e-04 13.312 1
9057 NUPR1 16_23 0.4846290 68.58 9.888e-05 -10.532 2
13435 DHRS11 17_22 0.5080183 61.81 9.342e-05 -8.142 1
7744 R3HCC1L 10_62 0.6724263 39.81 7.965e-05 7.439 1
12530 RP11-490G2.2 1_60 0.0008154 29860.63 7.245e-05 5.044 1Genes with largest z scores
genename region_tag susie_pip mu2 PVE z num_eqtl
7849 MC4R 18_33 0.264861 128.46 1.012e-04 13.312 1
7425 MST1R 3_35 0.000000 33940.06 0.000e+00 -12.621 2
38 RBM6 3_35 0.000000 39742.84 0.000e+00 12.536 1
9 SEMA3F 3_35 0.000000 38958.32 0.000e+00 12.511 1
7420 RNF123 3_35 0.000000 22473.32 0.000e+00 -10.959 1
1778 MAPK3 16_24 0.020311 98.48 5.951e-06 10.880 1
6171 TAOK2 16_24 0.023429 94.27 6.572e-06 10.738 1
8291 INO80E 16_24 0.023393 94.16 6.554e-06 10.734 1
9057 NUPR1 16_23 0.484629 68.58 9.888e-05 -10.532 2
10490 SULT1A1 16_23 0.048304 62.63 9.001e-06 10.415 1
11757 NPIPB7 16_23 0.024446 63.77 4.638e-06 10.038 1
7963 ZNF668 16_24 0.100919 77.13 2.316e-05 10.000 1
7964 ZNF646 16_24 0.100919 77.13 2.316e-05 -10.000 1
10597 SULT1A2 16_23 0.013171 56.47 2.213e-06 -9.958 2
12058 ETV5 3_114 0.893685 95.62 2.543e-04 9.862 1
5471 SAE1 19_33 0.004569 99.74 1.356e-06 9.849 1
7423 CAMKV 3_35 0.000000 51451.22 0.000e+00 -9.848 1
457 PRSS8 16_24 0.017222 72.05 3.692e-06 -9.765 1
11270 LAT 16_23 0.220743 56.62 3.719e-05 -9.553 1
2477 MTCH2 11_29 0.010422 83.55 2.591e-06 -9.551 1Comparing z scores and PIPs
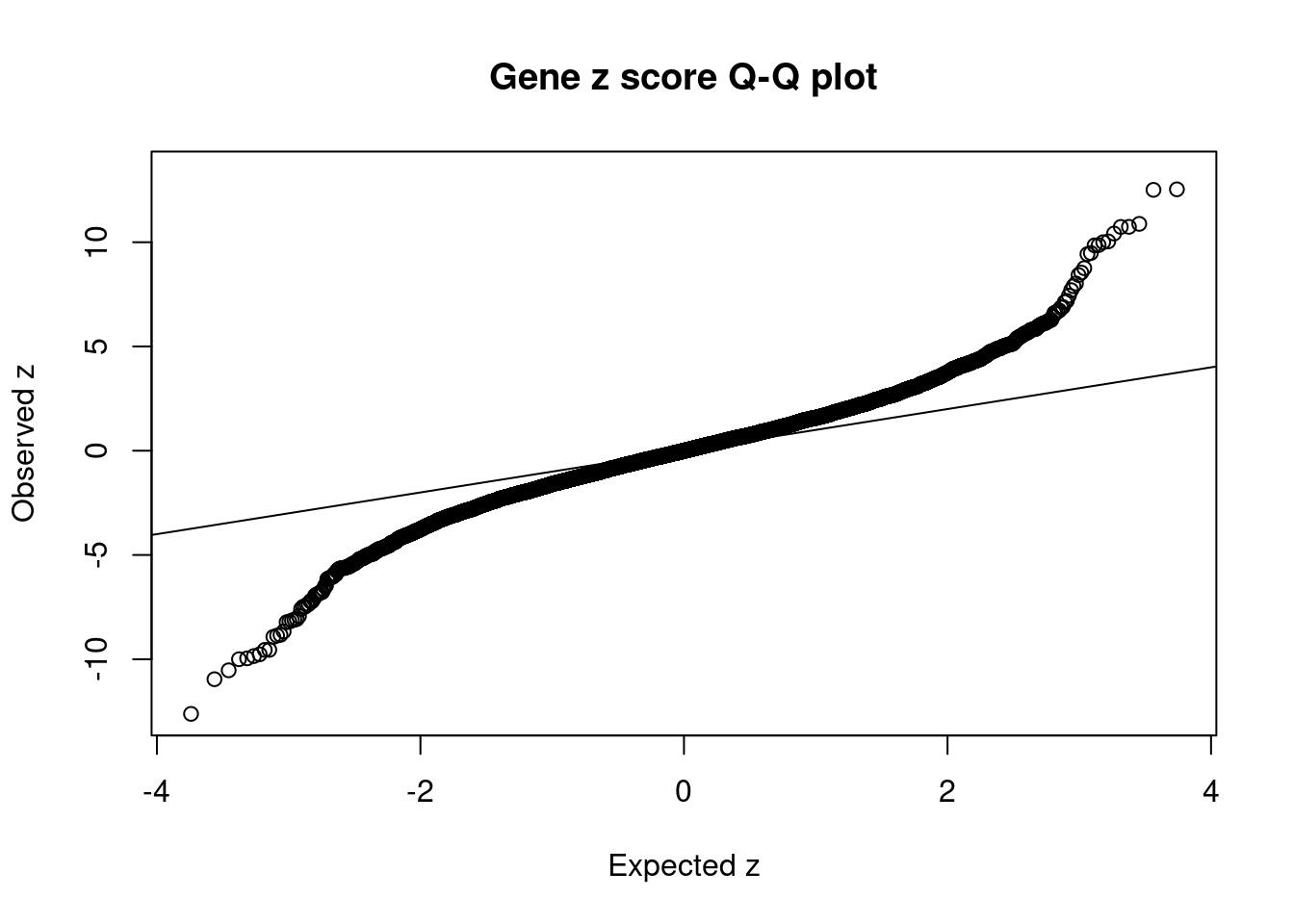
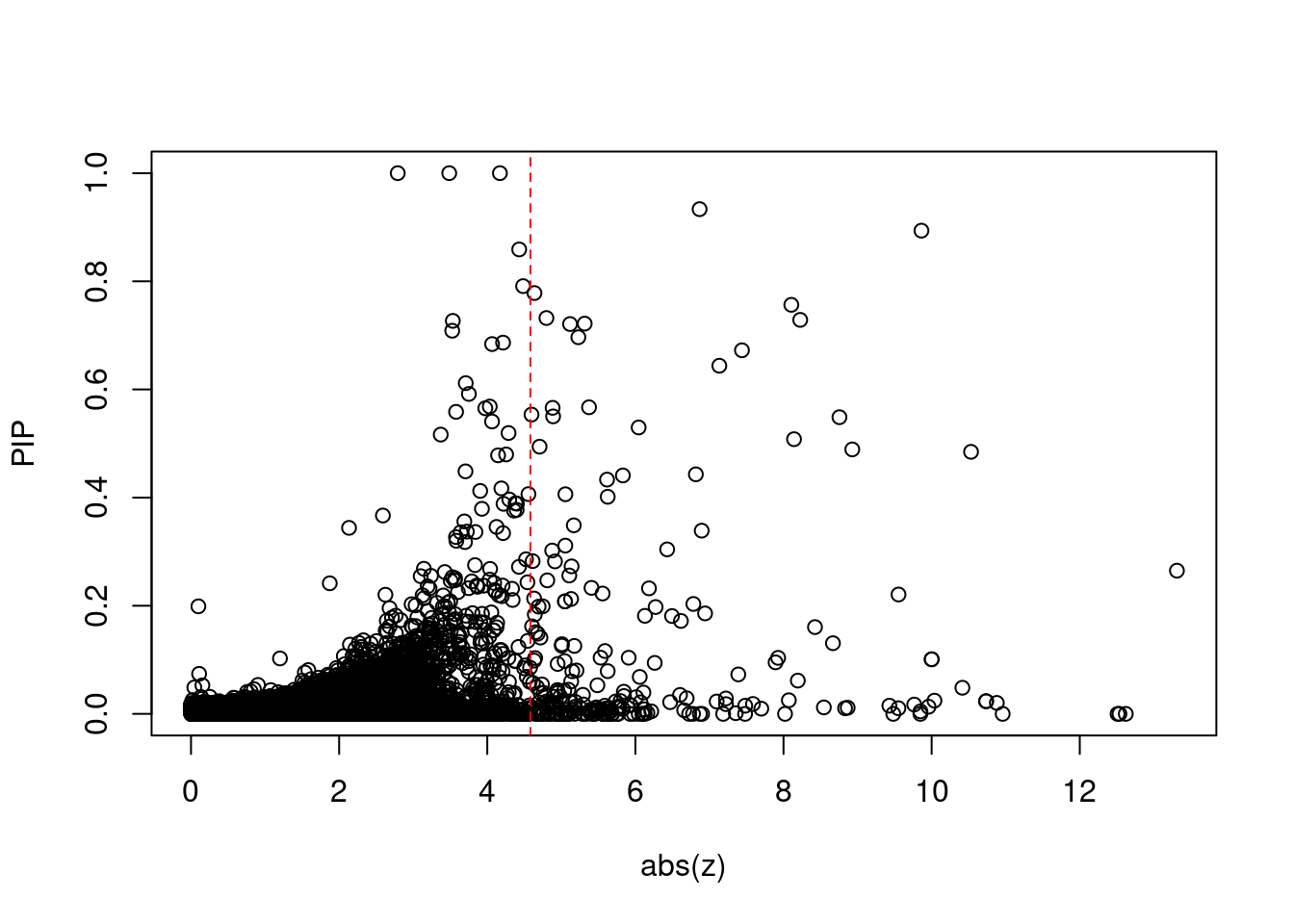
[1] 0.02272 genename region_tag susie_pip mu2 PVE z num_eqtl
7849 MC4R 18_33 0.264861 128.46 1.012e-04 13.312 1
7425 MST1R 3_35 0.000000 33940.06 0.000e+00 -12.621 2
38 RBM6 3_35 0.000000 39742.84 0.000e+00 12.536 1
9 SEMA3F 3_35 0.000000 38958.32 0.000e+00 12.511 1
7420 RNF123 3_35 0.000000 22473.32 0.000e+00 -10.959 1
1778 MAPK3 16_24 0.020311 98.48 5.951e-06 10.880 1
6171 TAOK2 16_24 0.023429 94.27 6.572e-06 10.738 1
8291 INO80E 16_24 0.023393 94.16 6.554e-06 10.734 1
9057 NUPR1 16_23 0.484629 68.58 9.888e-05 -10.532 2
10490 SULT1A1 16_23 0.048304 62.63 9.001e-06 10.415 1
11757 NPIPB7 16_23 0.024446 63.77 4.638e-06 10.038 1
7963 ZNF668 16_24 0.100919 77.13 2.316e-05 10.000 1
7964 ZNF646 16_24 0.100919 77.13 2.316e-05 -10.000 1
10597 SULT1A2 16_23 0.013171 56.47 2.213e-06 -9.958 2
12058 ETV5 3_114 0.893685 95.62 2.543e-04 9.862 1
5471 SAE1 19_33 0.004569 99.74 1.356e-06 9.849 1
7423 CAMKV 3_35 0.000000 51451.22 0.000e+00 -9.848 1
457 PRSS8 16_24 0.017222 72.05 3.692e-06 -9.765 1
11270 LAT 16_23 0.220743 56.62 3.719e-05 -9.553 1
2477 MTCH2 11_29 0.010422 83.55 2.591e-06 -9.551 1GO enrichment analysis for genes with PIP>0.5
#number of genes for gene set enrichment
length(genes)[1] 35Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"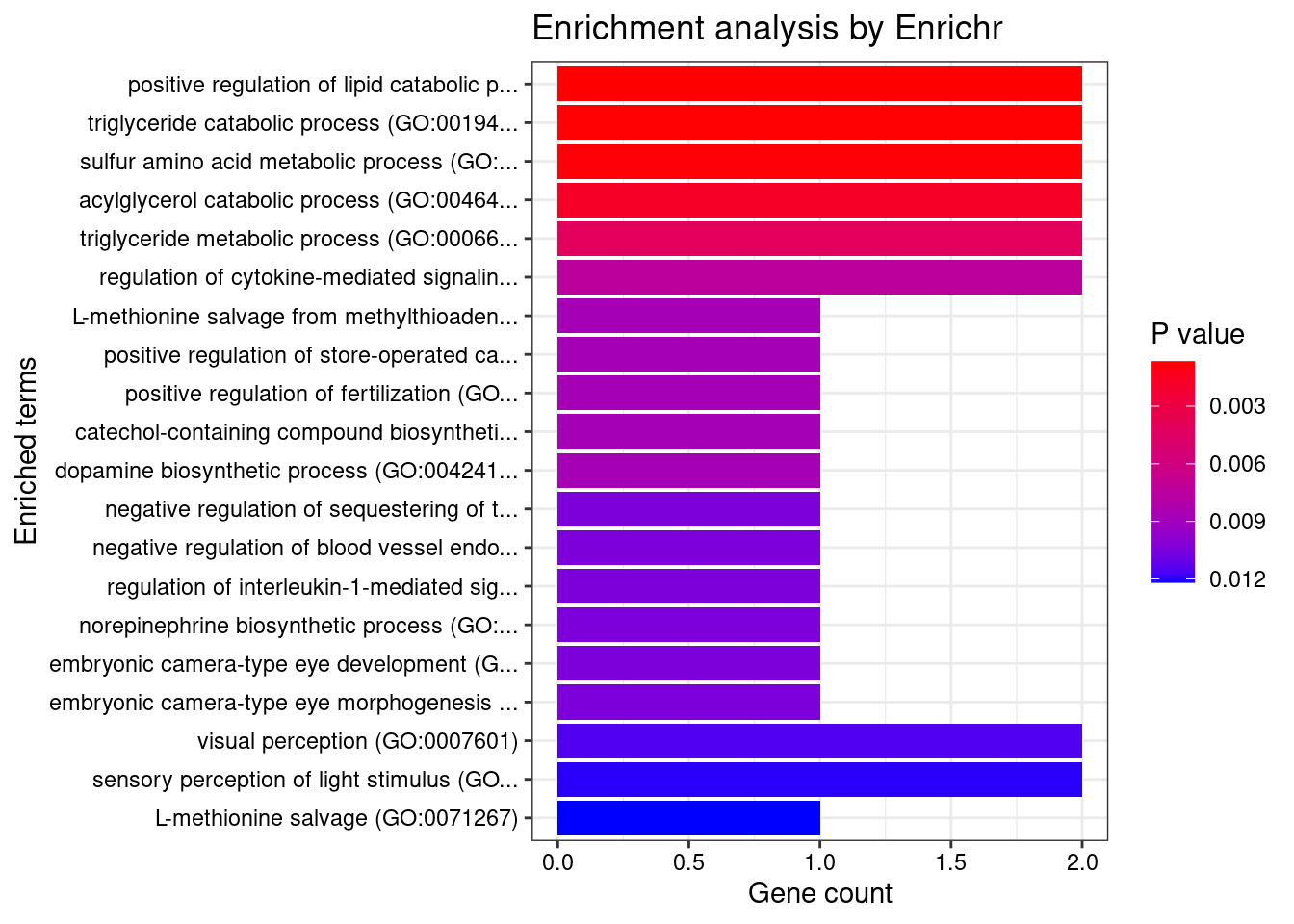
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"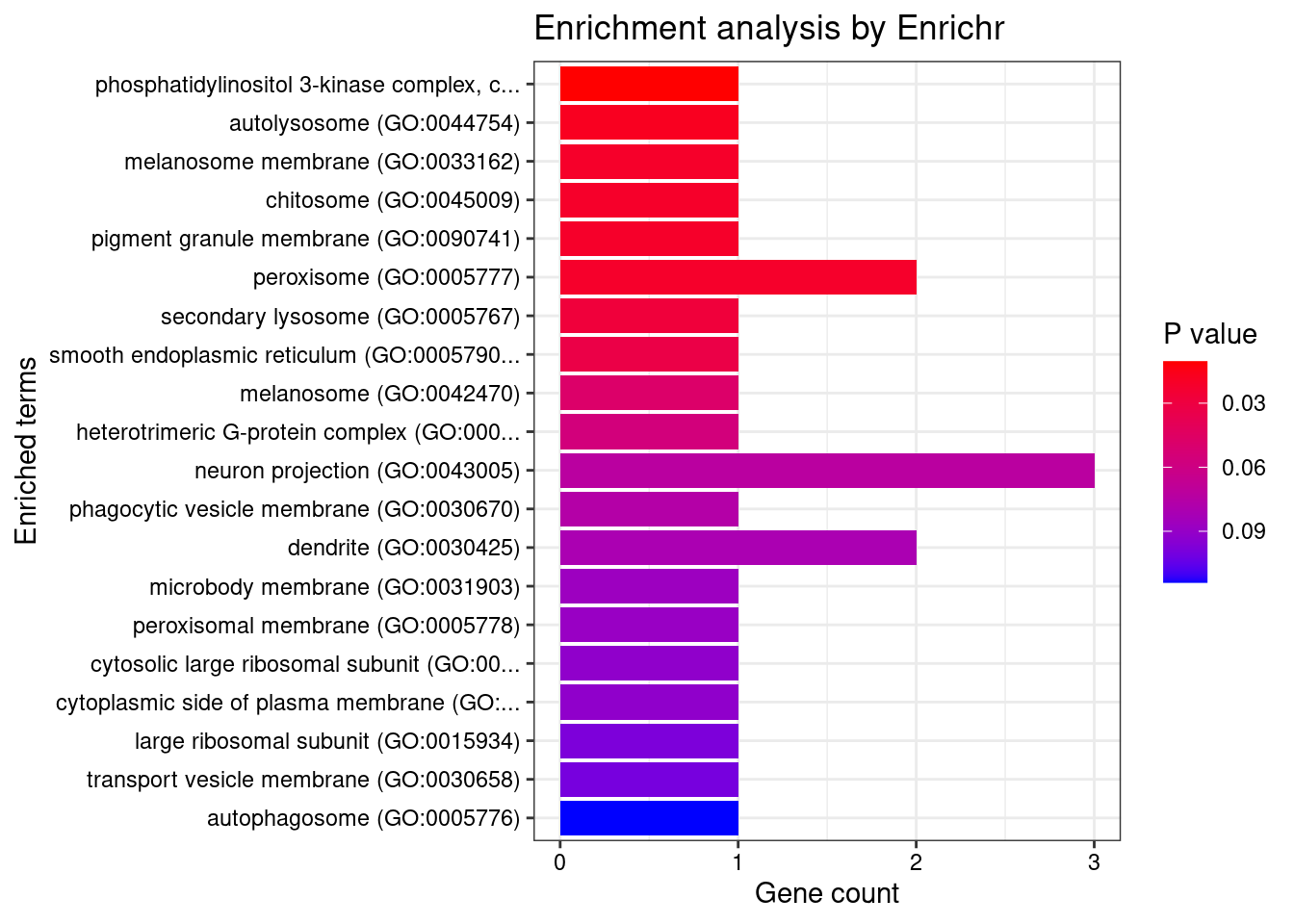
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)DisGeNET enrichment analysis for genes with PIP>0.5
Description
94 Carbamoyl-Phosphate Synthase I Deficiency Disease
112 OPTIC ATROPHY AND CATARACT, AUTOSOMAL DOMINANT
114 Neutral Lipid Storage Disease with Myopathy
115 Cerebral Cavernous Malformations 3
122 DYSTONIA, DOPA-RESPONSIVE, WITH OR WITHOUT HYPERPHENYLALANINEMIA, AUTOSOMAL RECESSIVE (disorder)
124 Familial cerebral cavernous malformation
128 CHARCOT-MARIE-TOOTH DISEASE, DOMINANT INTERMEDIATE F
131 PULMONARY HYPERTENSION, NEONATAL, SUSCEPTIBILITY TO
134 Hyperammonemia Due to Carbamoyl Phosphate Synthetase 1 Deficiency
135 Carbamoyl Phosphate Synthase 1 Deficiency
FDR Ratio BgRatio
94 0.02412 1/18 1/9703
112 0.02412 1/18 1/9703
114 0.02412 1/18 1/9703
115 0.02412 1/18 1/9703
122 0.02412 1/18 1/9703
124 0.02412 1/18 1/9703
128 0.02412 1/18 1/9703
131 0.02412 1/18 1/9703
134 0.02412 1/18 1/9703
135 0.02412 1/18 1/9703WebGestalt enrichment analysis for genes with PIP>0.5
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum =
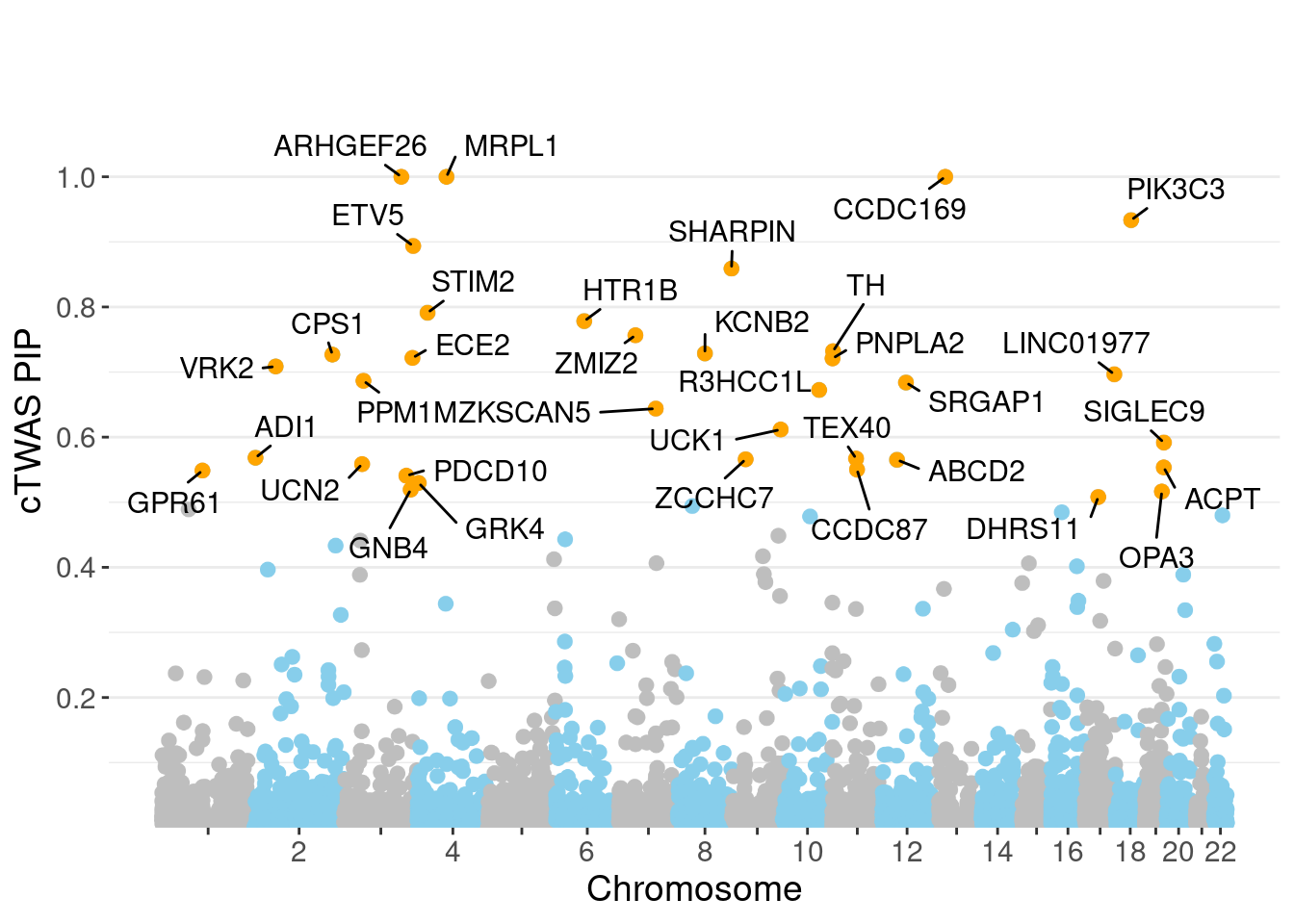
minNum, : No significant gene set is identified based on FDR 0.05!NULLPIP Manhattan Plot
Sensitivity, specificity and precision for silver standard genes
#number of genes in known annotations
print(length(known_annotations))[1] 41#number of genes in known annotations with imputed expression
print(sum(known_annotations %in% ctwas_gene_res$genename))[1] 24#significance threshold for TWAS
print(sig_thresh)[1] 4.583#number of ctwas genes
length(ctwas_genes)[1] 6#number of TWAS genes
length(twas_genes)[1] 248#show novel genes (ctwas genes with not in TWAS genes)
ctwas_gene_res[ctwas_gene_res$genename %in% novel_genes,report_cols] genename region_tag susie_pip mu2 PVE z num_eqtl
2884 ARHGEF26 3_95 1.0000 20272.04 6.031e-02 -4.171 1
8261 MRPL1 4_52 1.0000 18758.93 5.581e-02 3.486 1
9400 SHARPIN 8_94 0.8589 25.31 6.467e-05 -4.431 1
12013 CCDC169 13_13 1.0000 11401.83 3.392e-02 -2.791 1#sensitivity / recall
print(sensitivity) ctwas TWAS
0.00000 0.07317 #specificity
print(specificity) ctwas TWAS
0.9994 0.9775 #precision / PPV
print(precision) ctwas TWAS
0.0000 0.0121 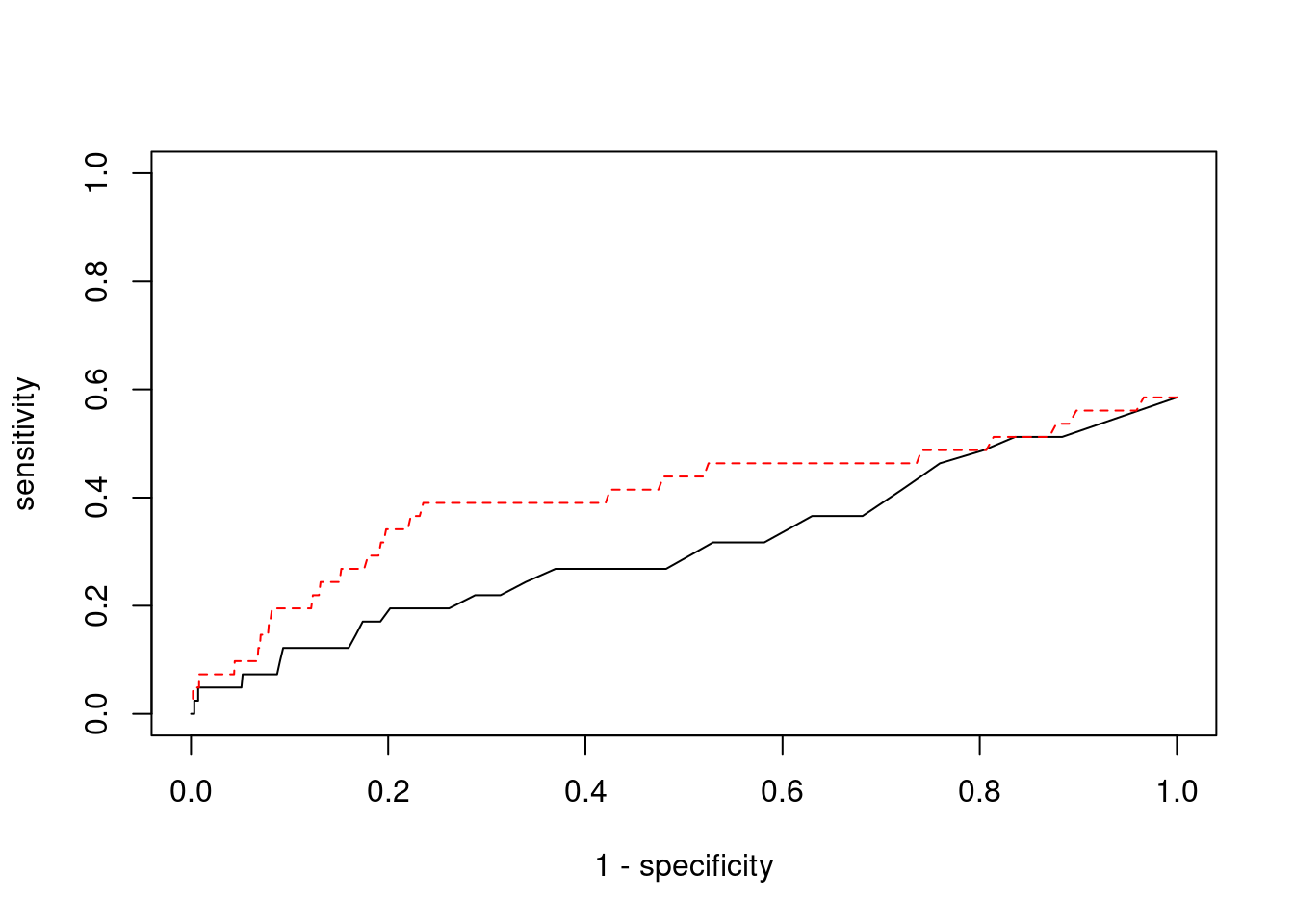
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] readxl_1.3.1 forcats_0.5.1 stringr_1.4.0 dplyr_1.0.7
[5] purrr_0.3.4 readr_2.1.1 tidyr_1.1.4 tidyverse_1.3.1
[9] tibble_3.1.6 WebGestaltR_0.4.4 disgenet2r_0.99.2 enrichR_3.0
[13] cowplot_1.0.0 ggplot2_3.3.5 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] fs_1.5.2 lubridate_1.8.0 bit64_4.0.5 doParallel_1.0.16
[5] httr_1.4.2 rprojroot_2.0.2 tools_3.6.1 backports_1.4.1
[9] doRNG_1.8.2 utf8_1.2.2 R6_2.5.1 vipor_0.4.5
[13] DBI_1.1.1 colorspace_2.0-2 withr_2.4.3 ggrastr_1.0.1
[17] tidyselect_1.1.1 bit_4.0.4 curl_4.3.2 compiler_3.6.1
[21] git2r_0.26.1 cli_3.1.0 rvest_1.0.2 Cairo_1.5-12.2
[25] xml2_1.3.3 labeling_0.4.2 scales_1.1.1 apcluster_1.4.8
[29] digest_0.6.29 rmarkdown_2.11 svglite_1.2.2 pkgconfig_2.0.3
[33] htmltools_0.5.2 dbplyr_2.1.1 fastmap_1.1.0 highr_0.9
[37] rlang_0.4.12 rstudioapi_0.13 RSQLite_2.2.8 jquerylib_0.1.4
[41] farver_2.1.0 generics_0.1.1 jsonlite_1.7.2 vroom_1.5.7
[45] magrittr_2.0.1 Matrix_1.2-18 ggbeeswarm_0.6.0 Rcpp_1.0.7
[49] munsell_0.5.0 fansi_0.5.0 gdtools_0.1.9 lifecycle_1.0.1
[53] stringi_1.7.6 whisker_0.3-2 yaml_2.2.1 plyr_1.8.6
[57] grid_3.6.1 blob_1.2.2 ggrepel_0.9.1 parallel_3.6.1
[61] promises_1.0.1 crayon_1.4.2 lattice_0.20-38 haven_2.4.3
[65] hms_1.1.1 knitr_1.36 pillar_1.6.4 igraph_1.2.10
[69] rjson_0.2.20 rngtools_1.5.2 reshape2_1.4.4 codetools_0.2-16
[73] reprex_2.0.1 glue_1.5.1 evaluate_0.14 data.table_1.14.2
[77] modelr_0.1.8 vctrs_0.3.8 tzdb_0.2.0 httpuv_1.5.1
[81] foreach_1.5.1 cellranger_1.1.0 gtable_0.3.0 assertthat_0.2.1
[85] cachem_1.0.6 xfun_0.29 broom_0.7.10 later_0.8.0
[89] iterators_1.0.13 beeswarm_0.2.3 memoise_2.0.1 ellipsis_0.3.2